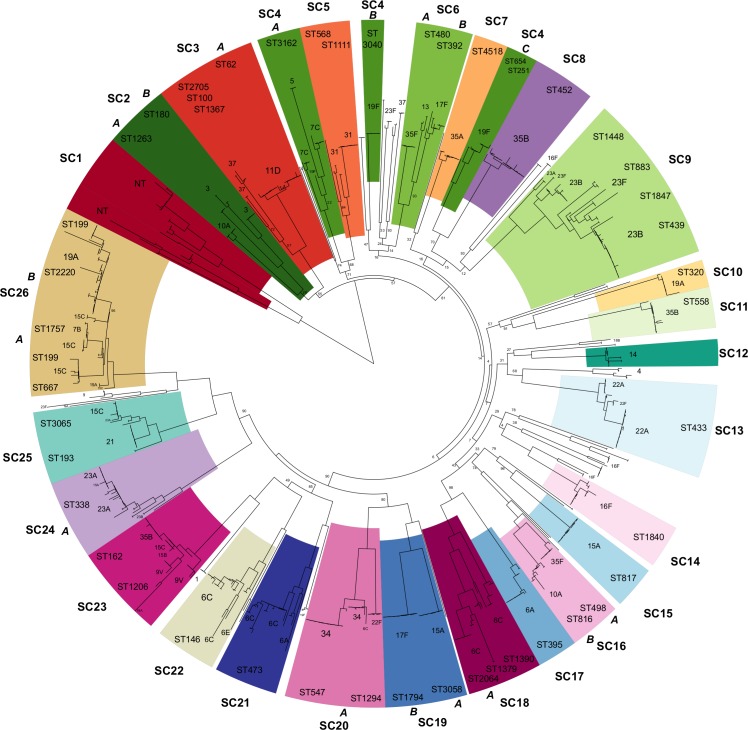
Fig 1. Phylogeny of pneumococcal population structure.
Maximum likelihood phylogeny of 937 pneumococcal carriage isolates inferred from an alignment of 1,111 core COGs including 78,525 polymorphic sites using RAxML with GTR+Γ nucleotide substitution model and 100 bootstrap replicates. Clades are colored by sequence cluster (SC), which are labeled on the outside ring. Some SCs are further divided into monophyletic sub-clusters (A, B, C) based on ancestral recombination history. Pneumococcal serotypes and MLST are labeled on each clade and bootstrap values are labeled on internal branches.