Author response image 1. Comparison of our in vivo vastus lateralis rhythmic dataset with mouse skeletal muscle rhythmic data (Dyar et al., 2014; Zhang et al., 2014).
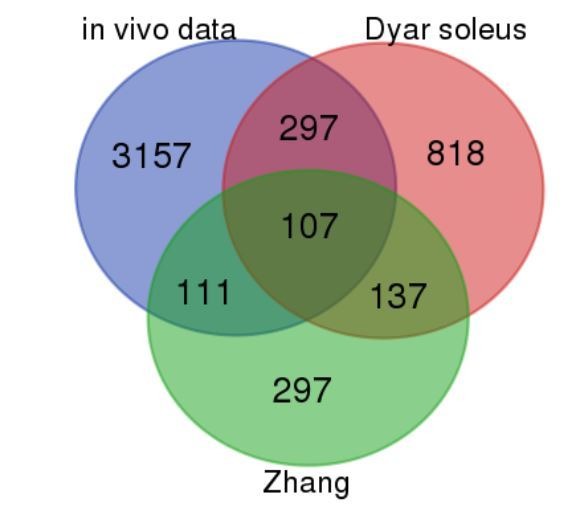
Blue = our in vivo RNAseq data; red = (Dyar et al., 2014) soleus; green = (Zhang et al., 2014). The numbers presented in the Venn diagram correspond to the total number of rhythmically expressed genes for each segment (exons only). For the human in vivo dataset (our work), this corresponds to R–E + R–I.R–E, with the cutoff described in the manuscript (Figure 1). For (Dyar et al., 2014) (microarray) and (Zhang et al., 2014) (microarray) rhythmic genes were identified using a Benjamini–Hochberg q–value <0.2 in the JTK_Cycle algorithm.
