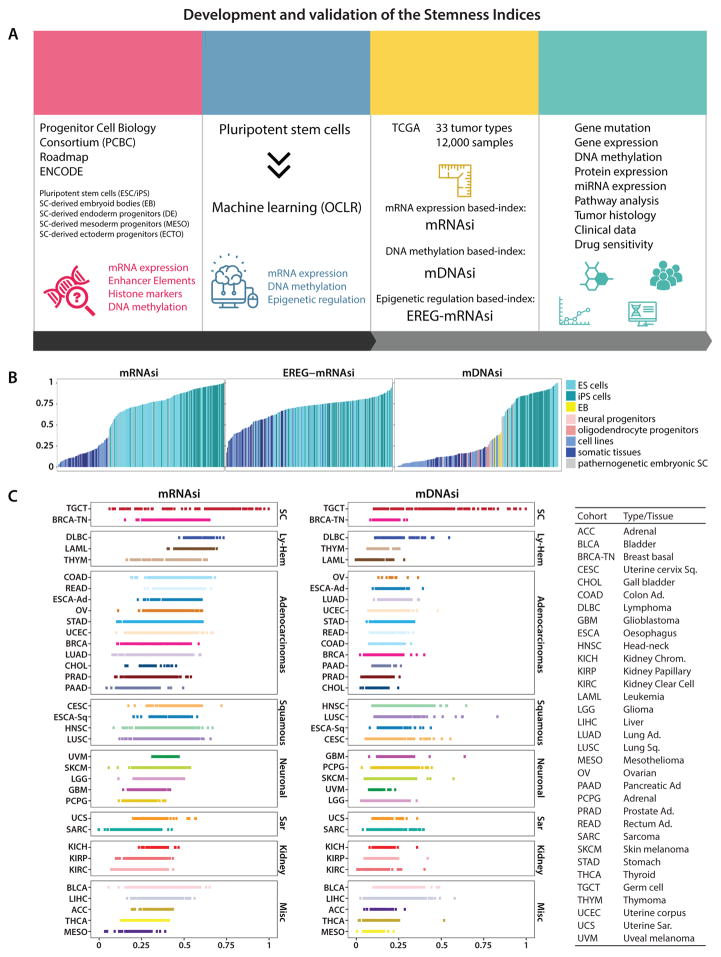
Figure 1. Development and validation of the Stemness Indices.
(A) Overall methodology. Highlighted are data sources Progenitor Cell Biology Consortium (PCBC), Roadmap and ENCODE databases, OCLR machine learning algorithm, and the resulting stemness indices mRNAsi, mDNAsi and EREG-mRNAsi. The indices for each TCGA tumor sample were correlated with known cancer biology, tumor pathology, clinical information, and drug sensitivity.
(B) Stemness indices of the validation set derived using our stemness signature.
(C) TCGA tumor types sorted by the stemness indices obtained from transcriptomic (mRNAsi) and epigenetic features (mDNAsi); indices were scaled from 0 (low) to 1 (high). The TCGA tumor types were grouped based on their histology and cell-of-origin into stem cell-like (SC), lympho-hematopoietic (Ly-Hem), Adenocarcinomas, Squamous Cell Carcinomas (Squamous), Neuronal lineage (Neuronal), Sarcomas (Sar), Kidney tumors (Kidney), and not belonging to any of the above (Misc) (Table S2).
See also Figures S1 and S2; and Tables S1 and S2.