Figure 12.
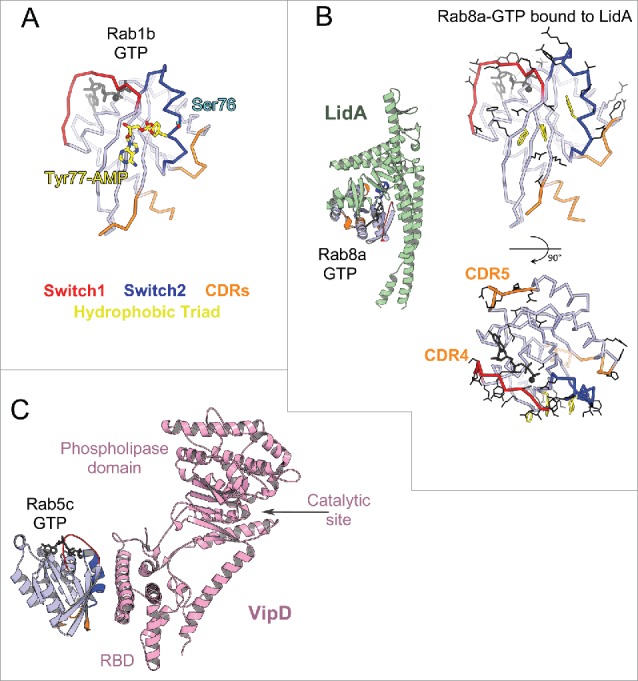
Bacterial Rab regulators. (A) Structure of active Rab1b modified by DrrA (3NKV). GTP analog and Mg2+ are shown in dark gray. The Tyr of Rab1b hydrophobic triad that is AMPylated (yellow) by DrrA is in the central part of the canonical partner binding site. The Rab1 Ser phosphocholination site in the Switch2 is also shown (cyan). (B) Legionella effector LidA bound to Rab8a (3TNF). Rab8a effector binding site (right) includes the canonical partner interaction surface (top) and expands to the adjacent surface (bottom). Rab8a residues changing solvent accessible area upon interaction with LidA are shown in black lines. (C) Legionella effector VipD bound to Rab5c (4KYI). Binding of Rab5c to VipD's helical RBD allosterically induces conformational changes in the phospholipase domain, resulting in opening of the catalytic site and activation of the enzymatic activity.
