Figure 1.
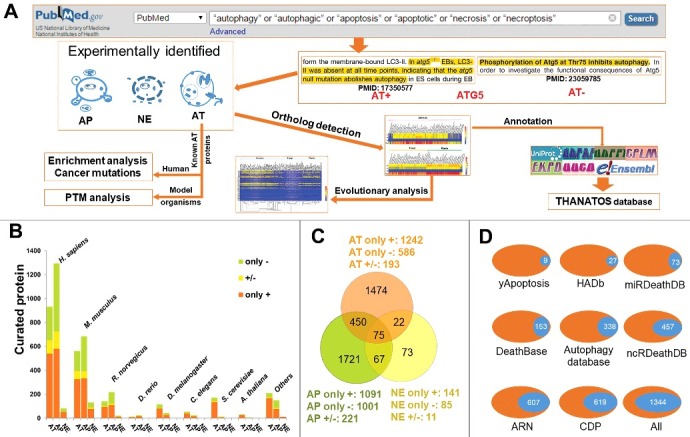
The collection and curation of proteins that were experimentally identified to be associated with autophagy and cell death pathways from the literature. (A) In this study, we used multiple keywords to search the PubMed search engine, and obtained a total of 4,237 known AT, AP and NE proteins. Using 3,882 known proteins from 8 model organisms, we computationally detected their potential orthologs in 164 eukaryotes, and further performed an evolutionary analysis of ATG genes. Also, we carried out the enrichment analysis and the cancer mutation analysis for known human AT proteins, while the PTM analysis was conducted for known AT proteins in model species. Finally, we combined both known and computationally identified AT, AP and NE proteins together and developed the THANATOS database. (B) Based on experimental evidence, we annotated each known protein with a “+” or “-” to distinguish the positive or negative regulation in autophagy or PCDs. For 3,882 known AT, AP and NE proteins of 8 organisms, the proteins annotated only with “+” (only +) or “-” (only -), and with both “+” and “-” (+/-) were separately present. (C) The overlap of different types of known proteins for 8 model species. (D) The comparison of curated proteins from the literature between THANATOS and other existing resources. i. All, the number of nonredundant proteins in 8 public databases.
