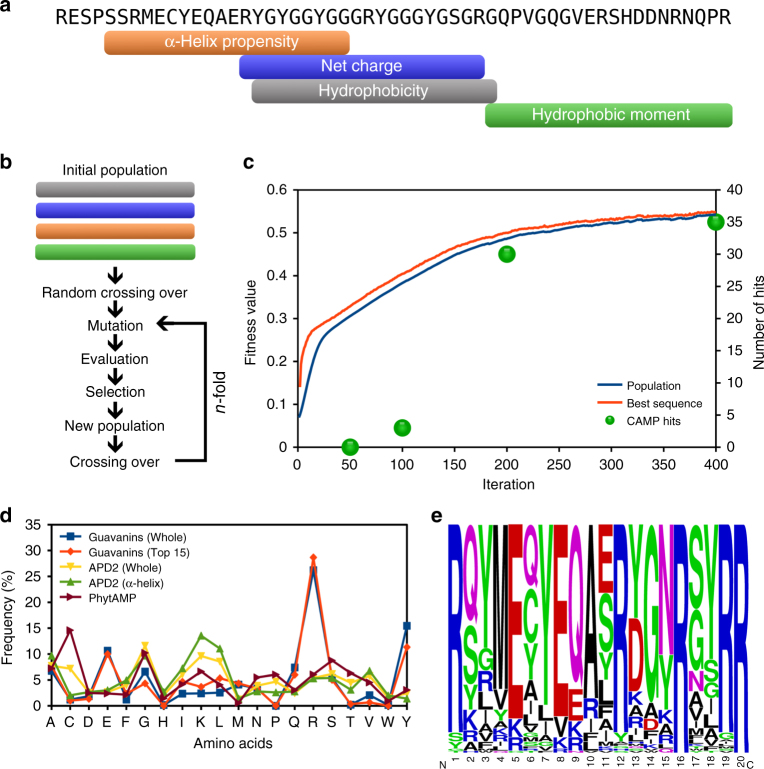
Fig. 1.
Design and selection of guavanins. a Fragment mapping into the Pg-AMP1 sequence. Each fragment represents the maximum value of its respective physicochemical property: in orange, the α-helix propensity (0.553); in blue, the positive net charge (+3); in gray, the average hydrophobicity (−0.092); and in green, the hydrophobic moment (0.3). b Flowchart of our custom genetic algorithm. The four Pg-AMP1 fragments were used as the initial population; in the first iteration a totally random sequence selection system for crossing over was applied, in order to improve the diversity of sequences and in the subsequent iterations a roulette wheel selection model was applied for selection of sequences for crossing over. c Fitness function evolution during the algorithm iterations and the number of CAMP hits of the highest ranked sequence at iterations 50, 100, 200, and 400; Guavanin sequences were retrieved from the 50th iteration. d Amino acid distribution of guavanins and AMPs from APD2 and PhytAMP. Blue squares represent data obtained from 100 guavanin sequences; orange diamonds, the top 15 guavanins; yellow down triangles, the overall APD2 composition; green up triangles, the composition of α-helical peptides from APD2; and brown right triangles the plant AMP sequences from PhytAMP (37). e The frequency logo of the 100 generated guavanin sequences (Supplementary Table 1), showing that they are arginine-rich peptides, Arg residues are in at least 20% of their compositions