Figure 1.
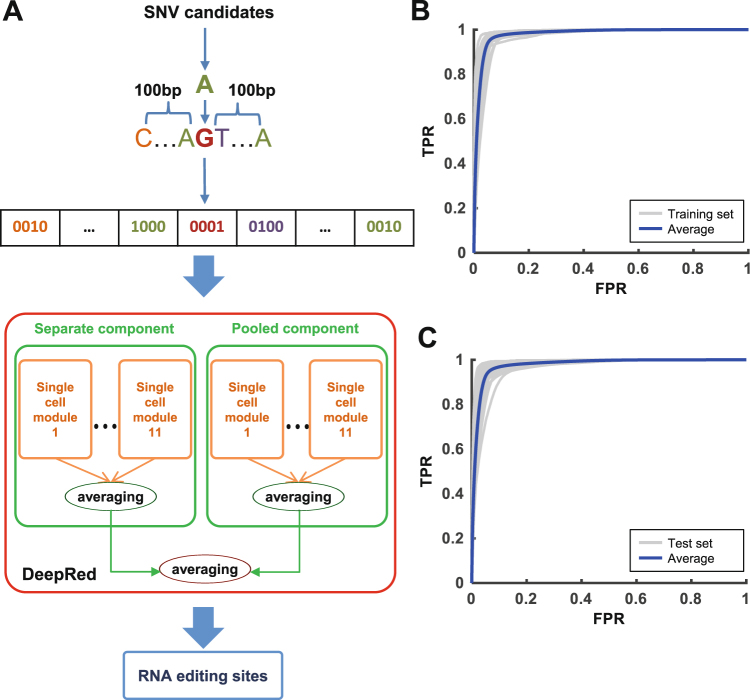
The architecture and performance of DeepRed. (A) The hybrid framework of DeepRed. DeepRed consists of separate and pooled components to account for the features of RNA editing derived from the separate samples and pooled samples methods, respectively. Each component combines 11 independent single-cell modules together using a simple averaging method. The separate and pooled components are combined by a simple averaging method. The input of DeepRed is a one-hot-encoded sequence of 201 base pairs (bp) centred at the candidate SNV. (B) The ROC curves (grey) of DeepRed were achieved in 22 gold standard sets in the training set. The average ROC curve of DeepRed in the training set is presented as a blue curve. (C) The ROC curves (grey) of DeepRed were achieved in 42 gold standard sets in the test set. The average ROC curve of DeepRed in the test set is presented as a blue curve.