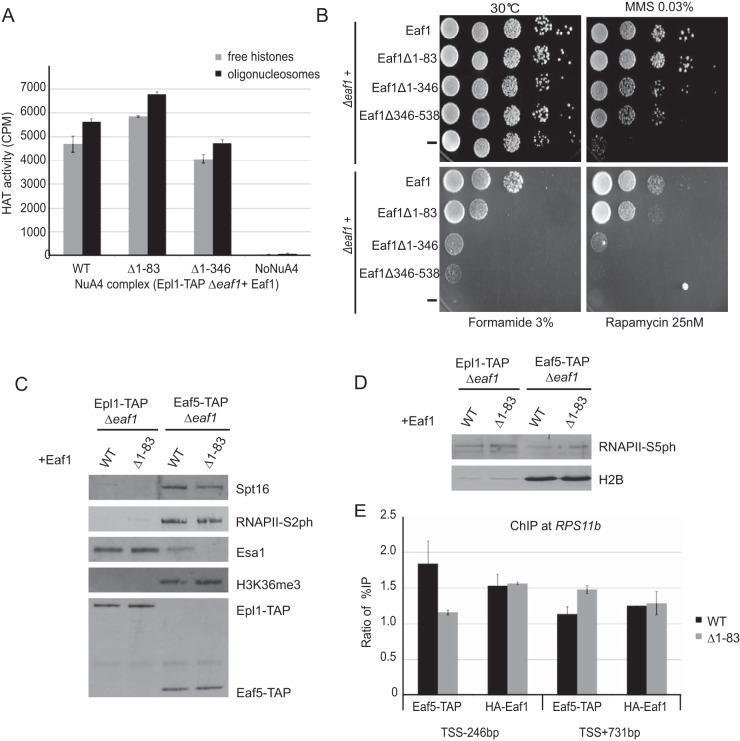
FIG 6.
Functional assays of Eaf1 mutants based on mapped molecular interactions. (A) The loss of TINTIN or Tra1 modules does not have an obvious impact on NuA4 HAT activity in vitro. NuA4 complexes purified through Epl1-TAP from cells expressing wild type or Eaf1 truncation mutants (Δ1-83 or Δ1-346) were tested in HAT assays using free histones or oligonucleosomes as the substrate with 3H-acetyl-CoA, followed by spotting on P81 membranes and scintillation counting. Error bars indicate the ranges of duplicate reactions. (B) Phenotypic analysis of yeast strains expressing wild type, different truncation mutants of Eaf1 (Δ1-83, Δ1-346, or Δ346-538) or a mutant expressing no Eaf1. Serial dilution spot assays were performed on rich medium alone or in the presence of formamide, the DNA-damaging agent methylmethane sulfonate (MMS), or the TOR pathway inhibitor rapamycin. Each mutant shows a specific growth phenotype. (C) Loss of association with NuA4 does not affect TINTIN interaction with elongating RNA polymerase on the body of genes (RNAPII-Ser2ph isoform), the FACT histone chaperone (Spt16), and the active coding region-enriched H3K36me3 histone mark. The tobacco etch virus nuclear inclusion a endopeptidase (TEV) elutions of IgG immunoprecipitations from cell extracts as in Fig. 5D (Epl1-TAP or Eaf5-TAP with wild type [WT] or Eaf1-Δ1-83) were analyzed by Western blotting with the indicated antibodies. (D) Loss of TINTIN does not disrupt NuA4 interaction with the early elongating polymerase at the 5′ ends of genes (RNAPII-Ser5ph isoform). Western blots of fractions as in panel C using RNAPII-Ser5ph and H2B antibodies are shown. (E) NuA4-dependent recruitment of TINTIN at the gene promoter but not on the coding region. eaf1 mutant cells (Δ1-83) show a loss of TINTIN (Eaf5-TAP) at the promoter of RPS11B gene but not on its coding region, whereas NuA4 recruitment (Eaf1) on the promoter is not affected by TINTIN. ChIP-qPCR was performed at the promoter region (TSS − 246 bp) and the coding region (TSS + 731 bp) of the NuA4 target gene RPS11B. The data represent the percent immunoprecipitation (IP)/input normalized on signal at the negative-control locus FMP27 (TSS + 5 kb). The error bars indicate the ranges between two biological replicates.