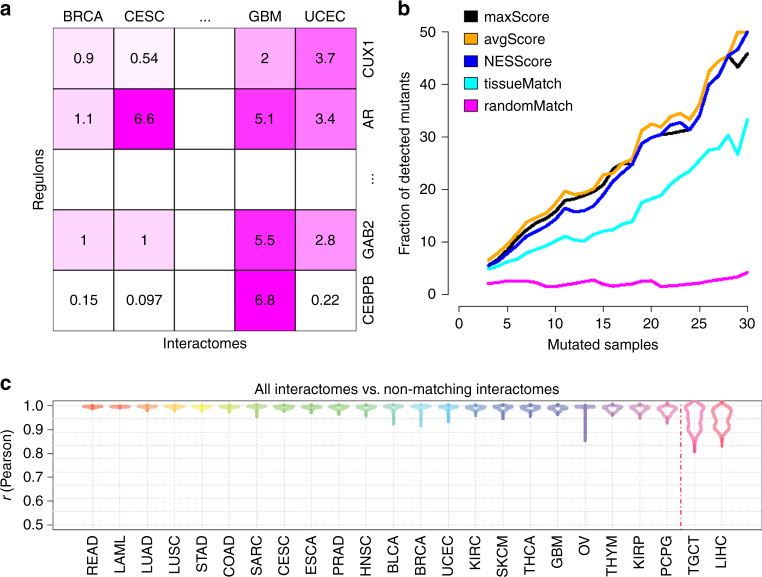
Fig. 1.
Inferring protein activity with metaVIPER. a Overview of metaVIPER. The set of transcriptional targets for each regulatory protein (its regulon) constitutes the fundamental building blocks of an interactome, which reflect its overall, context-specific regulatory control structure. MetaVIPER identifies the regulon that best recapitulates the regulatory targets of a protein by assessing its enrichment in the tissue-specific differential expression signature. In the example shown here, for instance, the regulon for protein CUX1 in an unknown or orphan tissue is better recapitulated by the uterine corpus endometrial carcinoma (UCEC)-based regulon, while the transcriptional program for the androgen receptor protein (AR) is better recapitulated by the cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) and glioblastoma (GBM)-based regulons. The numbers indicate –log10(p-value) for enrichment of the regulons on the gene expression signature, as computed by VIPER. b Impact of recurrent coding somatic mutations on metaVIPER-inferred protein activity. Fraction of proteins showing significant association between metaVIPER-inferred protein activity and somatic mutations (p < 0.01) is presented. VIPER analysis was performed using the tissue-matched network (tissueMatch), metaVIPER was performed by integrating the results from individual interactomes using maxScore, avgScore, and NESScore methods; the baseline control was computed by using intercatomes selected at random (randomMatch). The X-axis represents the minimum number of TCGA samples presenting the specific gene mutation required for inclusion of the encoded protein in the analysis. c Inference of protein activity for orphan tissues. MetaVIPER can effectively reproduce differential protein activity in TCGA tissues, even when the corresponding matched interactome is removed from the analysis. The only partial exception is represented by two tissue lineages—liver hepatocellular carcinoma (LIHC) and testicular germ cell tumors (TGCT)—which are defined by highly specific regulatory programs. The probability density distribution for the correlation between protein activities (NES) inferred by metaVIPER using all available interactomes vs. metaVIPER using all, but the tissue-matched interactome (Pearson’s correlation) across all samples is shown by the violin plots