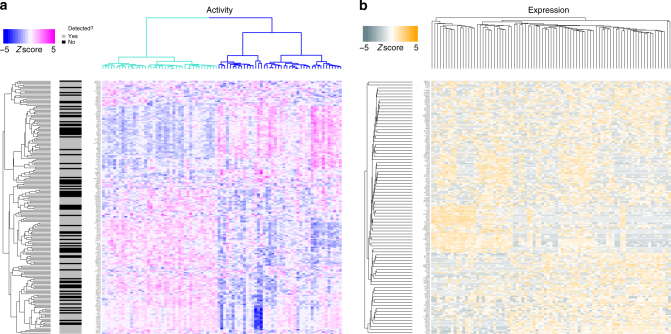
Fig. 2.
Inference of protein activity for single cells from GBM mouse model. a MetaVIPER-based protein activity analysis of single cells from a mouse GBM model27,28 by unsupervised clustering using all annotated transcriptional factors, co-transcriptional factors, and signaling proteins. Two major clusters were identified, corresponding to established mesenchymal (MES, blue) and proneural (PN, turquoise) subtypes, with varying proliferative (Prolif) potential11. Indeed, among the top 200 transcriptional factors (i.e., with the highest inter-cluster activity variability), we found established master regulatory transcriptional factors of the MES (FOSL1, FOSL2, RUNX1, CEBPB, CEBPD, MYCN, ELF4), PN (OLIG2, ZNF217), and Prolif (HMGB2, SMAD4, PTTG1, E2F1, E2F8, FOXM1) subtypes13. b Subtype representation is lost when clustering is performed based on gene expression profiles