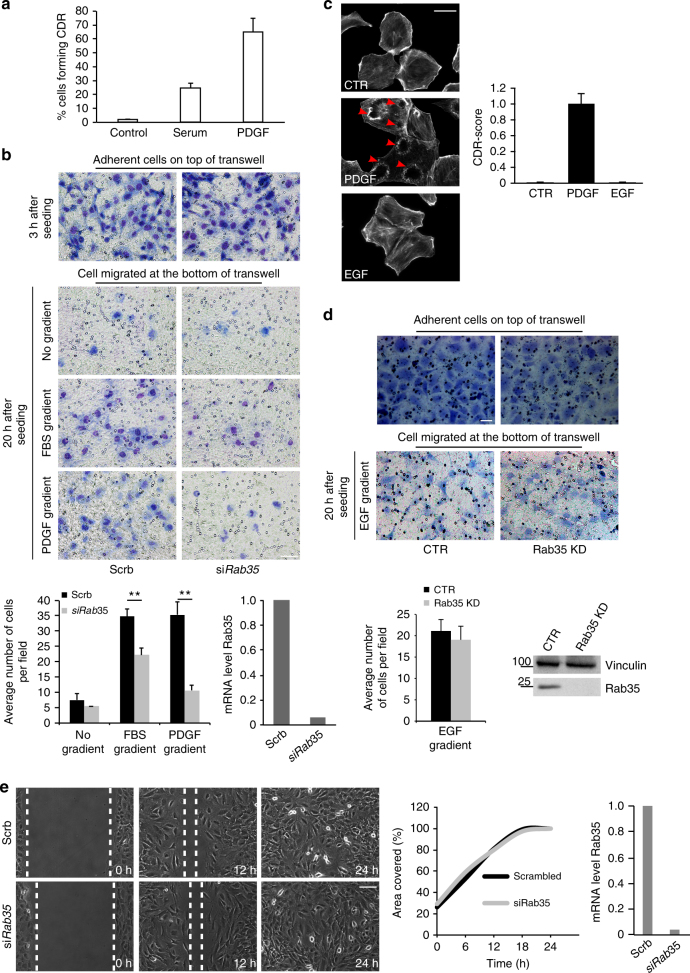
Fig. 5.
RAB35 is dispensable for chemotaxis when CDR do not form. a Serum-starved MEF were stimulated with PDGF (20 ng/ml) or 10% serum for 10’, fixed and stained with FITC-Phalloidin to detect F-actin and CDRs. We plotted % cell forming CDRs. Data are the mean ± SD (n = 120 cells/experiments, three independent ones). b Scrambled (Scrb) and Rab35-silenced (siRab35) MEFs seeded into 8-μm pore Transwell with PDGF (20 ng/ml) or 10% serum added in the lower chamber. Crystal violet staining was done after 3 h to detect total number of seeded cells, and after 20 h to detect migrated cells to the lower side. Chemotaxis was quantified as number of cells/field ± SD (>6 fields of view/condition, in two independent experiments). Data are the mean ± SD. **p < 0.01. Gene silencing was verified by qRTPCR. Scale bar, 50 μm. c Serum-starved MEFs were stimulated with PDGF (20 ng/ml) or EGF (100 ng/ml) for 10′, fixed and stained with FITC-Phalloidin to detect F-actin and CDRs. Red arrows indicate CDRs. CDR score was calculated by normalizing the number of CDR-positive cells per each condition against the PDGF-stimulated sample, used as control. Data are the mean ± SD (n > 100 cells/condition in two independent experiments). Scale Bar, 20 μm. d Scrambled (CTR) and Rab35-silenced (Rab35 KD) MEFs were seeded into 8-μm pore Transwell. PDGF (20 ng/ml) or EGF (100 ng/ml) was added to the lower chamber. Cells migrated through the transwell were stained counted as described in b. Scale bar, 40 μm. The extent of chemotaxis was quantified as the average number of cells/field ± SD (>10 fields of view/condition, in two independent experiments). Data are the mean ± SD, **p < 0.01. RAB35 downregulation was assessed by immunoblotting. Vinculin is a loading control. e Confluent scrambled (Scrb) and Rab35-silenced (siRab35) MEFs were scratched with a pipette tip and recorded by time-lapse. Still images at the indicated time point are shown (Supplementary Movie 9). The percentage of free area covered was determined with ImageJ tool. A representative experiment out of four is shown. Gene silencing was verified by qRTPCR. Scale Bar, 100 μm