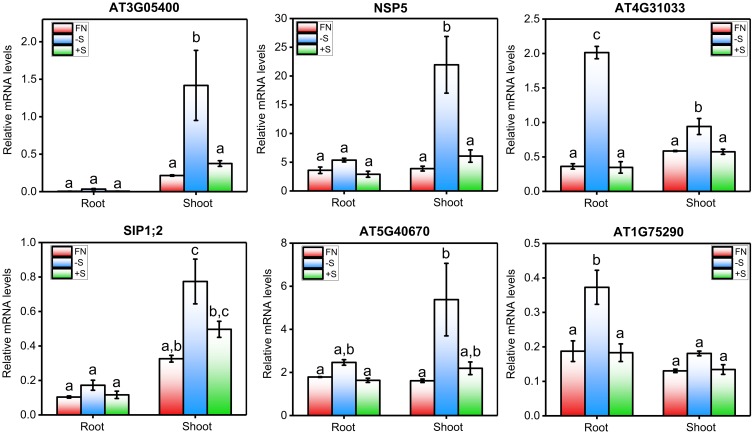
FIGURE 2.
qPCR-analyzed expression profiles of highly consistent genes whose function in the sulfate response is unknown. mRNA levels of six genes (AT3G05400, NSP5, AT4G31033, SIP1;2, AT5G40670, and AT1G75290) selected from the meta-analysis of the sulfate transcriptome data were analyzed by qPCR using the clathrin adapter gene (AT4G24550) as a normalization reference gene. Briefly, 5-day-old seedlings were transferred to liquid MS medium that was supplemented with sulfate (FN, 1.5 mM S) or not, after which the seedlings were allowed to grow for 2 days. At dawn on the 7th day, the plants were treated with potassium sulfate (+S, 1.5 mM) or potassium chloride (–S, 3 mM). The mRNA levels were measured 2 h after treatment using the total RNA extracted from root and shoot samples. The values plotted correspond to the means of three independent experiments ± standard errors of the mean; 80 to 100 seedlings were used per replicate. The means with different lowercase letters are significantly different, p < 0.05 (one-way ANOVA and Tukey’s test).