FIGURE 2.
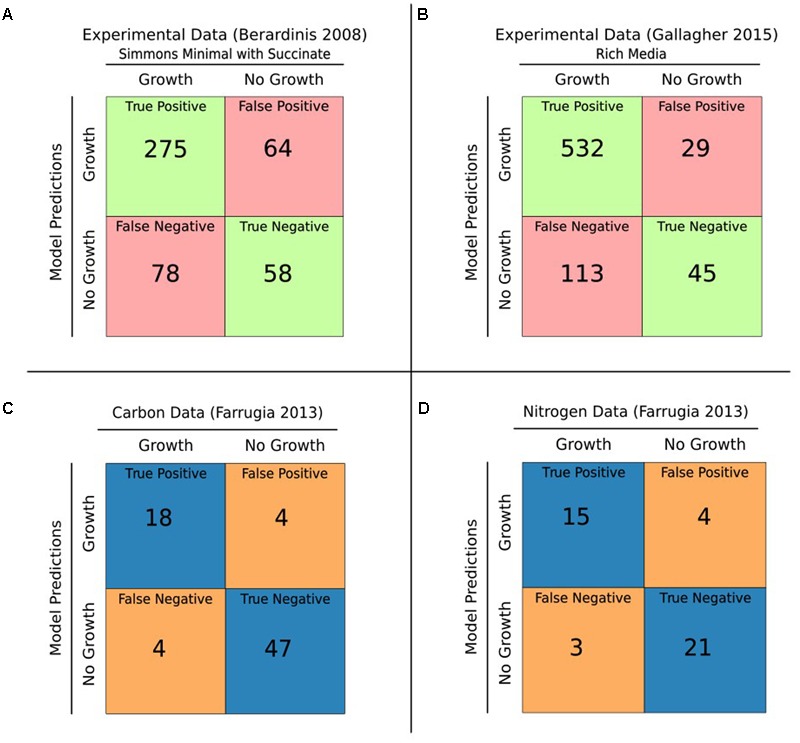
Gene essentiality and growth predictions. (A) iCN718 was used to predict gene essentiality. The results were compared to the de Berardinis et al. (2008) experimental dataset with 68% accuracy. (B) iCN718 predicted gene essentiality results compared with the Gallagher et al. (2015) dataset exhibited 80% accuracy. It is worth noting that the Berardinis dataset was of Acinetobacter baylyi ADP1 and therefore not every gene in iCN718 had an orthologous gene in the essentiality dataset. Green represents correct predictions, red represents incorrect predictions. The Gallagher dataset is from Acinetobacter baumannii strain AB5075 of which there is an ortholog for every gene within iCN718. Model-predicted ability to catabolize various sole carbon (C) and sole nitrogen (D) sources compared to the Farrugia et al. (2013) Biolog Phenotypic Array data for Acinetobacter baumannii AYE exhibited 89% and 84% accuracy, respectively. Blue represents correct predictions, orange represents incorrect predictions. Only compounds readily mapped to model metabolites were included from the Biolog data.
