Figure 1.
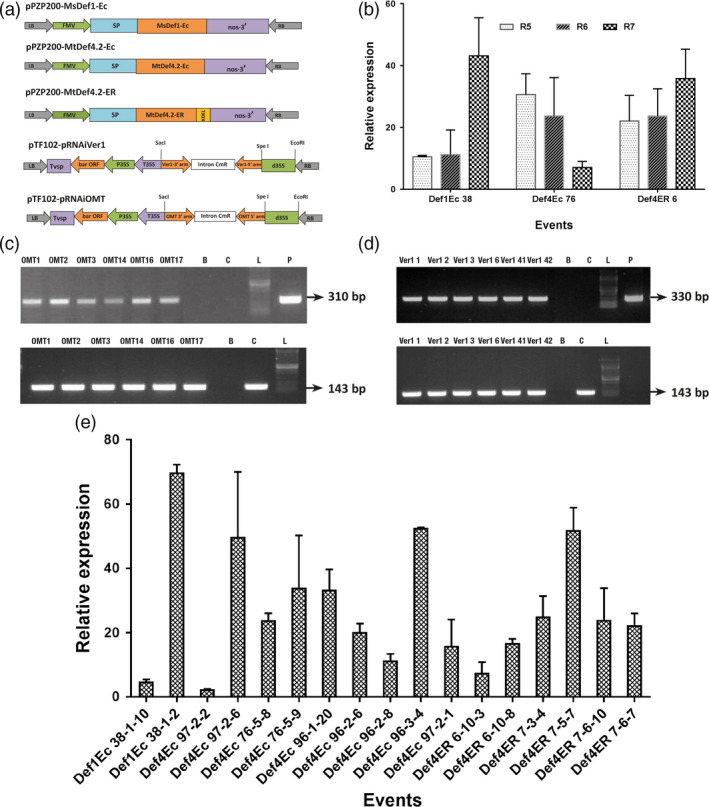
Transformation vectors and expression analysis of peanut OE‐Def and HIGS lines. (a) Expression vectors used for peanut transformation. The constitutive figwort mosaic virus (FMV) 35S promoter was used for expression of full‐length MsDef1‐Ec, MtDef4.2‐Ec and MtDef4.2‐ER . MsDef1‐Ec and MtDef4.2‐EC constructs targeted each defensin to the apoplast with signal peptide, whereas MtDef4.2‐ER construct retained this defensin in the endoplasmic reticulum. For targeting the aflatoxin pathway genes, the hpRNA cassettes had inverted repeats of respective omtA (aflP) and ver‐1(aflM) regions around the PR10 intron under the control of double CaMV 35S promoter. LB, left border; RB, right border; nos, nopaline synthase gene terminator; CaMV35S, cauliflower mosaic virus promoter; SP, signal peptide. (b) Expression of defensin transgenes in various OE‐Def events (pooled across generations) at different pod development stages (R5, R6 and R7). (c,d) RT‐PCR analyses to detect the expression of hpRNA transcripts in
mature cotyledons. A 310‐bp amplicon for omtA (c) and a 330‐bp amplicon for ver‐1 (d). An intron‐spanning peanut ADH3 gene was used as a control (lower panel). A 400‐bp amplicon is expected from a genomic DNA template, whereas 143‐bp amplicon is expected from a cDNA template. Letters B and C represent Blank and WT control, respectively; L stands for marker ladder and P denotes plasmid. (e) Expression of defensin transgenes in various OE‐Def events (pooled across generations) in mature cotyledons after infection with A. flavus AF11‐4 at 72 hpi. The housekeeping gene, G6PD was used for normalization with respect to the WT. Error bars represent the standard error (SE) of at least five replicates.
