Figure 2.
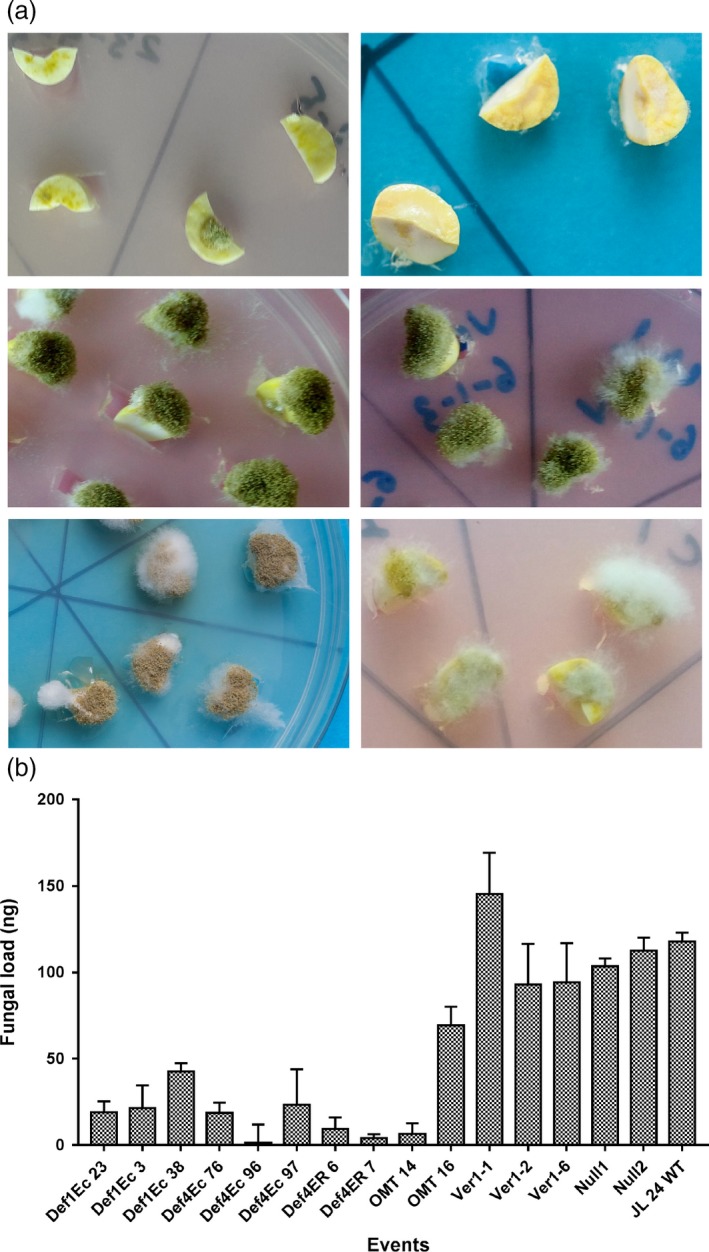
Fungal assay of OE‐Def and HIGS lines at 72 hpi. (a) Comparison of fungal colonization on cotyledons of MtDef4‐Ec 96 (top row left), MsDef1‐Ec 23 (top row right), HIGS line; hp‐omtA 16 (middle row left), HIGS lines; hp‐ver1‐1 6 (middle row right), WT control (last row left) and resistant check, 55‐437 (last row right); OE‐Def lines show no or very little fungal growth on events generated with extracellularly targeted Def4 and Def1 genes; HIGS lines show no restriction to fungal growth on events generated with omtA and ver‐1; extensive fungal growth and sporulation on WT controls, resistant check‐peanut variety 55‐437. (b) Fungal load of A. flavus on cotyledons of OE‐Def, HIGS and WT lines after 72 hpi. Error bar represents standard error of at least three biological replicates at P = 0.5.
