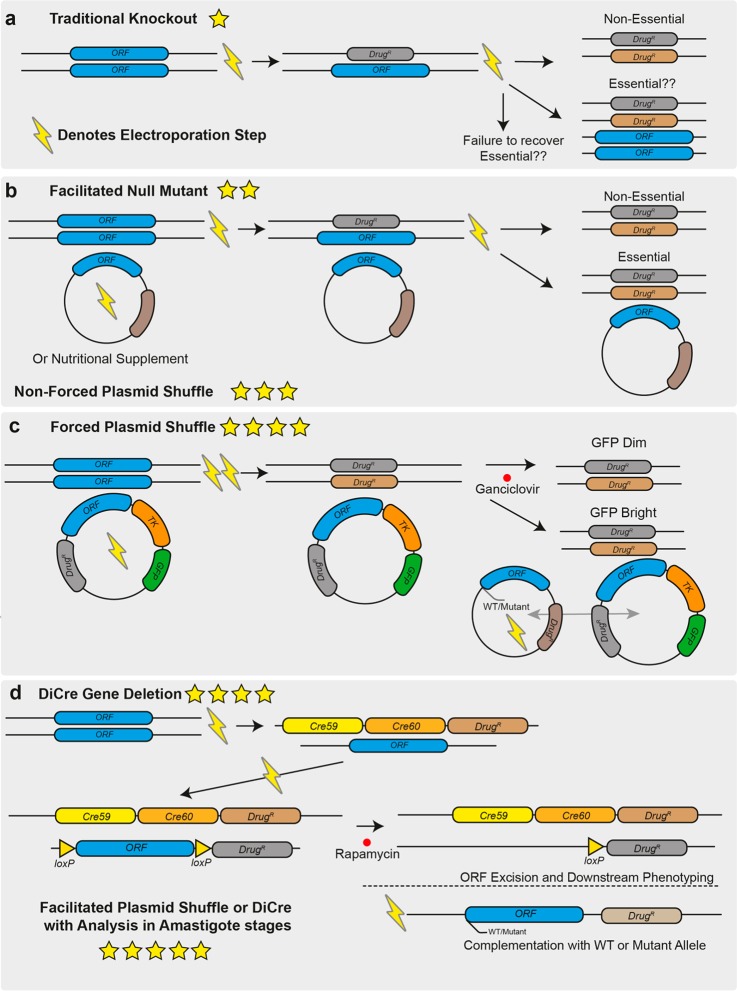
Figure 1.
Overview of techniques that can be used for genetic target validation in Leishmania. (a) Gene deletion by homologous replacement. Drug resistance markers are targeted to the gene of interest by long homology flanks (0.5–1 kb) in sequential transfections by electroporation. This process can now be facilitated using CRISPR/Cas9 and short homology flanked cassettes in a single transfection. Deletions targeting essential genes will result in cell death and failure to isolate null mutants or in ploidy changes that allow the cell to retain alleles of the wild-type locus as well as drug resistance markers. (b) Facilitated null mutant with unforced plasmid shuffle. An episome expressing the gene of interest is first transfected into the cell line or a nutritional supplement is provided, allowing it to survive the subsequent deletion of the chromosomal alleles of the gene of interest. The drug selection pressure for the episome can be removed and retention of the plasmid can be determined if the gene is not essential; then it will be possible to isolate parasites that lack the episome. (c) Forced plasmid shuffle. As in B, an episome expressing the gene of interest is transfected into the parasite to allow the deletion of the chromosomal alleles of the target gene. The episome also encodes a negative-selectable marker, herpes simplex virus thymidine kinase. Selection with ganciclovir favors the survival of parasites that lack the episome, so if a gene is nonessential, the episome will rapidly be lost from the population but will be retained for an essential gene despite the associated costs. The addition of a second episome containing mutant versions of the gene of interest allows for exploration of the roles of specific domains and residues in the encoded protein for correct gene function by assessing which plasmid of the two is preferentially retained. (d) DiCre inducible gene deletion. One allele of the target gene is replaced by a drug-selectable cassette containing a “floxed” allele, and in a second transfection stage, the remaining allele is replaced by a second drug resistance marker. The addition of rapamycin induces DiCre dimerization and excision of the floxed allele, and the phenotypes that emerge in the induced null mutants can then be analyzed. As in C, complementation allows for the assessment of null mutant specificity and functional assessment of defined domains or residues in the protein. In all panels, the number of stars indicate the quality of the genetic evidence for gene essentiality, with one star being the weakest and five stars being the strongest.