Fig. 4.
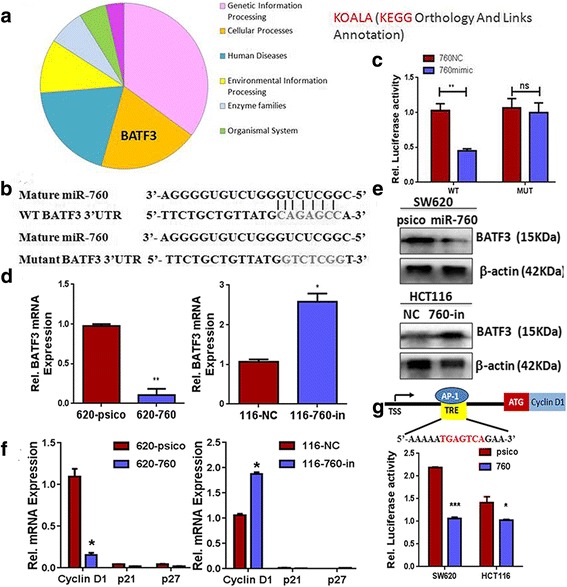
miR-760 directly targeted the 3′-UTR of BATF3 mRNA. a Annotation of predicted top 50 target genes using KOALA (http://www.kegg.jp/blastkoala/). BATF3 was revealed as a potential target related to cellular processes. b Schematic representation of miR-760 target sites in the 3′-UTR of BATF3 mRNA and a miR-760 mutant containing seven altered nucleotides in the seed sequence (mutant BATF33′-UTR). c Luciferase assay of pmir-GLO reporter plasmids carrying wild-type or mutant BATF3 3′-UTR co-transfected with miR-760 mimic (50 nM, 760 mimic) or negative control (50 nM, 760NC) in 293 T cells. Expression of BATF3 mRNA (d) and protein (e) in SW620 (d left and e upper) and HCT116 cells (d right and e bottom) transfected with the indicated miRNAs. f Real-time PCR analysis of cyclinD1, p21, and p27 mRNA expression in SW620 cells (left) and HCT116 (right). g (upper) The AP-1 binding site is located at 1.1 kb downstream of the cyclin D1 transcription start site (TSS). (bottom) miR-760-overexpressed SW620 or HCT116 cells were transfected with the wild-type (WT) cyclin D1 promoter-luciferase reporter plasmids and expression was assessed by the luciferase assay. Error bar represents the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001
