Abstract
The RecQ helicases play important roles in genome maintenance and DNA metabolism (replication, recombination, repair, and transcription). Five different homologs are present in humans, three of which are implicated in accelerated aging genetic disorders: Rothmund Thomson (RECQL4), Werner (WRN), and Bloom (BLM) syndromes. While the DNA helicase activities of the 5 human RecQ helicases have been extensively characterized, much less is known about their DNA double strand annealing activities. Strand annealing is an important integral enzymatic activity in DNA metabolism, including DNA repair. Here, we have characterized the strand annealing activities of all five human RecQ helicase proteins and compared them. Interestingly, the relative strand annealing activities of the five RecQ proteins are not directly (inversely) related to their helicase activities. RECQL5 possesses relatively strong annealing activity on long or small duplexed substrates compared to the other RecQs. Additionally, the strand annealing activity of RECQL5 is not inhibited by the presence of ATP, unlike the other RecQs. We also show that RECQL5 efficiently catalyzes annealing of RNA to DNA in vitro in the presence or absence of ATP, revealing a possible new function for RECQL5. Additionally, we investigate how different known RecQ interacting proteins, RPA, Ku, FEN1 and RAD51, regulate their strand annealing activity. Collectively, we find that the human RecQ proteins possess differential DNA double strand annealing activities and we speculate on their individual roles in DNA repair. This insight is important in view of the many cellular DNA metabolic actions of the RecQ proteins and elucidates their unique functions in the cell.
Graphical Abstract
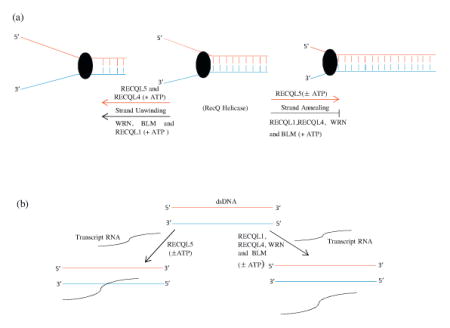
A model of RecQ helicases function to strand unwinding or strand annealing. A model of RecQ helicases function to strand unwinding or strand annealing. (a) (
 ) represent weak helicase activity (RECQL5 and RECQL4) and (←) represent strong helicase activity (WRN, BLM and RECQL1). (
) represent weak helicase activity (RECQL5 and RECQL4) and (←) represent strong helicase activity (WRN, BLM and RECQL1). (
 ) represent strong strand annealing activity (RECQL5) in the presence or absence of ATP and (
) represent strong strand annealing activity (RECQL5) in the presence or absence of ATP and (
 ) represent inhibition of strand annealing activity (RECQL1, RECQL1, WRN and BLM) in the presence of ATP (see Table 2). (b) RECQL5 promotes annealing of RNA to dsDNA in the presence or absence of ATP whereas remaining four RecQ helicases could not anneal RNA to dsDNA (±ATP). (
) represent inhibition of strand annealing activity (RECQL1, RECQL1, WRN and BLM) in the presence of ATP (see Table 2). (b) RECQL5 promotes annealing of RNA to dsDNA in the presence or absence of ATP whereas remaining four RecQ helicases could not anneal RNA to dsDNA (±ATP). (
 ) represent RecQ helicase.
) represent RecQ helicase.
1. Introduction
Helicases are ubiquitous enzymes that bind and remodel nucleic acids (DNA or RNA) [1–3]. They are essential during DNA replication and repair [4,5]. They unwind DNA using the energy source in ATP [4] and are classified according to their amino acids and helicase domain sequences into superfamilies [1,2,6,7]. RecQ helicases are DNA helicases that translocate along DNA strands in the 3′–5′ direction [8,9]. They have been shown to be particularly important in genome maintenance and are called guardians of the genome. RecQ helicases can unwind different oligonucleotide based DNA substrates e.g. fork duplexes, Holliday junctions and G-quadruplexes [3,10–12]. Many RecQ helicases are upregulated in cancer and transformed cells [3,4,8,10,13].
Defects in three of the five known human RECQ homologues are associated with different autosomal recessive disorders characterized by genomic instability and cancer predisposition [3,14]. Mutations in BLM cause Bloom syndrome, mutation in WRN cause Werner syndrome and mutations in RECQL4 cause Rothmund-Thomson syndrome, RAPADILINO syndrome and Baller–Gerold syndrome [4,15,16]. No genetic disorders have as yet been identified with RECQL1 or RECQL5 mutations, although RECQL1 and RECQL5 have important roles in cancer and aging [17–19]. RECQL5 participates in cell proliferation and transcription [20–22]. RECQL5 is an important elongation factor for preserving genome stability during transcription. [23,24]. Thus, RECQL5 depletion causes transcriptional stress (increase in the average rate, increased stalling, arrest, and backtracking) that leads to genome instability in the cell [20,22]. RECQL5 deficient mice are cancer prone [25,26]. This suggests that RECQL5 may act as a tumor suppressor, exhibiting elevated frequencies of spontaneous DNA double-strand breaks and homologous recombination (HR).
DNA strand annealing is known as heteroduplex formation between two complementary single stranded DNA or RNA molecules. Strand annealing may occur spontaneously, but it is promoted in vivo by certain annealing proteins. Strand annealing activity has been reported for the human RecQ helicases, and while this enzymatic process may be as important as the helicase activity, it has not yet been the focus of many studies. Strand annealing plays important roles in DNA metabolism including stabilization of replication forks, telomere (D-loop and T-loop structure) regulation, chromatin remodeling, DNA double strand break (DSB) repair and regulation of transcription [27]. There are two major pathways that repair DNA DSBs, homologous recombination (HR) and non-homologous end joining (NHEJ) [28]. HR repairs DSB using a template DNA strand or homologous sequences so that the original DNA sequence is conserved (error free) and specifically occurs during S and G2 phases of the cell cycle whereas NHEJ directly joins the two ends of a DSB regardless of DNA template sequence, is error prone and occurs throughout the cell cycle. The HR pathways occur sequentially starting with binding of HR factors at the DSB site, DNA end resection, formation of a D-loop or Holliday junction and processing of the Holliday junction or single strand annealing synthesis dependent pathway [29]. The NHEJ pathway exits in two forms, canonical NHEJ (C-NHEJ) and alternative NHEJ (Alt-NHEJ) [28]. The c-NHEJ pathway is initiated by recruitment of the Ku70/80 heterodimer, and then different c-NHEJ processing proteins tailor the DNA ends to permit ligation of the DSB DNA ends. Alt-NHEJ involves greater DNA DSB resection and repair at the site using small regions of microhomology at the ends of the DNA strand [28]. Thus, strand annealing is an error-prone recombination process that relies exclusively on annealing reaction intermediates to generate a heteroduplex DNA molecule.
A longstanding goal of our work has been to gain insight into the unique and combined functions of the five human RecQ proteins. In the yeast Saccharomyces cerevisiae and other simple organisms there is only one RecQ helicase and it is therefore of interest to understand why humans have five of these proteins. Are they redundant, or do they have complementary functions? How do they collaborate? In the pursuit of these questions we have addressed their pathway involvement by different approaches including analysis of their individual functions and their functional protein partners. While there is a large body of literature regarding the relative roles of these proteins as helicases, it will be interesting to explore the strand annealing activity of RecQs in a biological contexts [13].
Here, we characterize the annealing activity of all the human RecQs. We found that RECQL5 has relatively strong annealing activity independent of ATP, whereas the strand annealing activity of the other four human RecQs were inhibited by ATP. The impact on strand annealing of known RecQ interacting proteins such as RPA, Ku, FEN1, and Rad51for each human RecQ protein was also evaluated. Our findings highlight major differences between RECQL5’s strand annealing activity and that of the other four human RecQs. In addition, we found a novel function for RECQL5 as it catalyzes annealing of RNA to DNA in the presence or absence of ATP. Interestingly, the annealing of RNA to DNA activity was not found in the other RecQ proteins and this study is the first to report that RECQL5 has the unique function of catalyzing the annealing of RNA to DNA. While we usually think of the RecQ family of proteins as helicases, many biological functions attributed to these proteins are not dependent on their helicase activity. Thus, further understanding of the biochemical importance of strand annealing and how the annealing might function in maintenance of the genome integrity and regulation of DNA double strand break repair is clearly needed.
2. Materials and methods
2.1. Proteins expression and purification
Recombinant RECQL5 protein was purified from Escherichia coli by overexpression as a fusion protein with a self-cleaving intein–chitin-binding domain in BL21(DE3) CodonPlus RIPL strain (Stratagene, Agilent Technologies, Santa Clara, CA, USA), as previously described [30]. Recombinant His-tagged RECQL1 protein was overexpressed using a baculovirus/Sf9 insect system as previously described [31]. Recombinant histidine-tagged WRN protein was purified using a baculovirus/insect cell expression system, as previously described [32]. Recombinant RECQL4 with a C-terminal 9-histidine tag in the pGEX6p1 vector (GE Healthcare) was expressed and purified from E. coli Rosetta2 (DE3) (Novagen, EMD Millipore) as described previously [33]. Recombinant BLM was purified as described previously [30]. Recombinant histidine-tagged FEN1 protein was purified from E. coli, as previously described [34]. Recombinant RPA and RAD51 proteins were purified as described previously [35,36]. The recombinant human Ku 70/80 heterodimer was kindly provided by Dale Ramsden (University of North Carolina, Chapel Hill, NC).
2.2. DNA substrates
Oligonucleotides were synthesized and PAGE purified by Integrated DNA Technologies (Coralville, IA), and their sequences, in 5′–3′ orientation, are listed in Table 1 [33]. The oligomers were radiolabeled on the 5′ end with [γ-32P] ATP (3000Ci/mmol) with polynucleotide kinase (Roche Biochemical) per manufacturer’s directions and unincorporated nucleotides were removed using MicroSpin G-25 columns (GE Healthcare). To anneal the fork and full duplex substrates, oligonucleotides were combined in annealing buffer (40 mM Tris-HCL pH 8.0 and 50 mM NaCl) at a 1:2 ratio (labelled to unlabeled oligonucleotide), heated to 80 °C for 5 min, then cooled gradually to room temperature. Substrates used for individual experiments are indicated in the figure legends and depicted in each figure.
Table 1.
Oligonucleotides used in this study.
| Structure | Substrate | Sequence (5′–3′) |
|---|---|---|
|
(a/b) | T1 GGAATTCTACCAGTGCCTTGCTAGGACATCTTTGCCCA |
| 22/15(fork-1) | B1 CTAGACAGCTCCATGTAGCAAGGCACTGGTAGAATTC | |
|
|
Blunt end 22 base pairs | T2 GAGTGTGGTGTACATGCACTAC |
| B2 GTAGTGCATGTACACCACACTC | ||
|
|
Blunt end 80 base pairs | T3 GCTGATCAACCCTACATGTGTAGGTAACCCTAACCCTAACCCT |
| AAGGACAACCCTAGTGAAGCTTGTAACCCTAGGAGCT | ||
| B3 AGCTCCTAGGGTTACAAGCTTCACTAGGGTTGTCCTTAGGGTT | ||
| AGGGTTAGGGTTACCTACACATGTAGGGTTGATCAGC | ||
|
|
dsDNA 32/53 base pairs | T4 TTATTGTCTCATTAGCGGATACATATTTGAAT |
| B4 ATTCAAATATGTATCCGCTAATGAGACAATAACCCTG | ||
| ATAAATGCTTCACTAG | ||
|
|
ssDNA | T5 GAAGCATTTATCAGGGTTATTGTCTCATGAGCGGA |
| TACATATTTGAAT | ||
|
|
ssRNA | T6 GAAGCAUUUAUCAGGGUUAUUGUCUCAUGAGCGGA |
| UACAUAUUUGAAU |
2.3. DNA strand annealing
The DNA strand annealing activity of RECQL1, WRN, BLM, RECQL4, and RECQL5 were measured using single stranded complementary synthetic oligonucleotides (0.5 nM) of 80-mers or 22-mers in length (see Table 1, T3 and B3 make up the 80-mers and T2 and B2 make up the 22-mers) with the B strands labeled on the 5′ end using [γ-32P] ATP using T4 polynucleotide kinase. Annealing reactions (10 μl) were carried out in a time dependent manner (0, 1, 2, 5, 10, 15, and 30 min) at 37 °C in buffer (30 mM Tris–HCl pH 7.5, 50 mM KCl, 1 mM DTT, 5 mM MgCl2, BSA 100 μg/ml, plus or minus 5 mM ATP, when indicated), the reaction was started with the addition of B strand. Similarly, annealing reactions for each RecQ protein (each 10 nM) with increasing amounts of RPA, Ku, FEN1, and Rad51 as indicated were carried out at 37 °C in buffer (30 mM Tris–HCl pH 7.5, 50 mM KCl, 1 mM DTT, 5 mM MgCl2, BSA 100 μg/ml,), without ATP in the reactions, for 10 min. Each of the interacting proteins (RPA, Ku, FEN1 and Rad51) were added before the RecQ proteins. Reactions were stopped by addition of stop buffer (30 mM EDTA, 0.9% SDS, 30 mM Tris-Cl pH 8.0, 30% glycerol, 0.05% bromophenol blue, and 0.05% xylene cyanol). The reaction products were separated by electrophoresis on a 10% native poly-acrylamide gel in 1 × Tris-Borate-EDTA buffer (1X TBE) at 200 V for 1 h, then exposed to a PhosphorImager screen (GE Healthcare, Piscataway, NJ), and imaged on a Typhoon scanner (GE Healthcare, Piscataway, NJ). ImageQuanTL was employed to analyze and calculate the percentage of free and annealed substrate in each reaction. Each lane was normalized against background. Assays were done at least in triplicate, and representative gels are shown.
2.4. DNA–DNA and RNA–DNA annealing
DNA–DNA and RNA–DNA annealing was done as described previously with slight modification [22]. All oligonucleotides sequences (T4, B4, T5 and T6) used are shown in Table 1. Tailed dsDNA (T4 and B4) (each 0.5 nM) was incubated in the presence of RECQL5 (10 nM) in a buffer containing 30 mM Tris–HCl pH 7.5, 50 mM KCl, 1 mM DTT, 5 mM MgCl2, BSA 100 μg/ml for 0, 1, 2, 5, 10, 15 and 30 min at 37 °C. Annealing reactions were initiated by adding [γ-32P] labelled ssRNA (T6) or ssDNA (T5) (0.5 nM). Aliquots were withdrawn at the indicated time points and deproteinized by incubating samples in stop solution and analyzed as described under DNA strand annealing.
2.5. Helicase assay
Helicase activity of RECQL1, WRN, BLM, RECQL4, and RECQL5 (at concentrations mentioned in the figure legends) were measured using complementary synthetic oligonucleotides fork-1 duplex, shown in Table 1 (0.5 nM) for 30 min at 37 °C in a reaction volume of 10 μl reaction buffer containing 30 mM Tris–HCl pH 7.4, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 100 μg/ml BSA, 10% glycerol, 2.5 mM ATP. Where specified 10 nM unlabeled single stranded oligonucleotide was also present. Unless otherwise indicated, helicase reactions were terminated by the addition of 10 μl of stop buffer and analyzed as described under DNA strand annealing.
3. Results
3.1. RecQ proteins promote DNA strand annealing
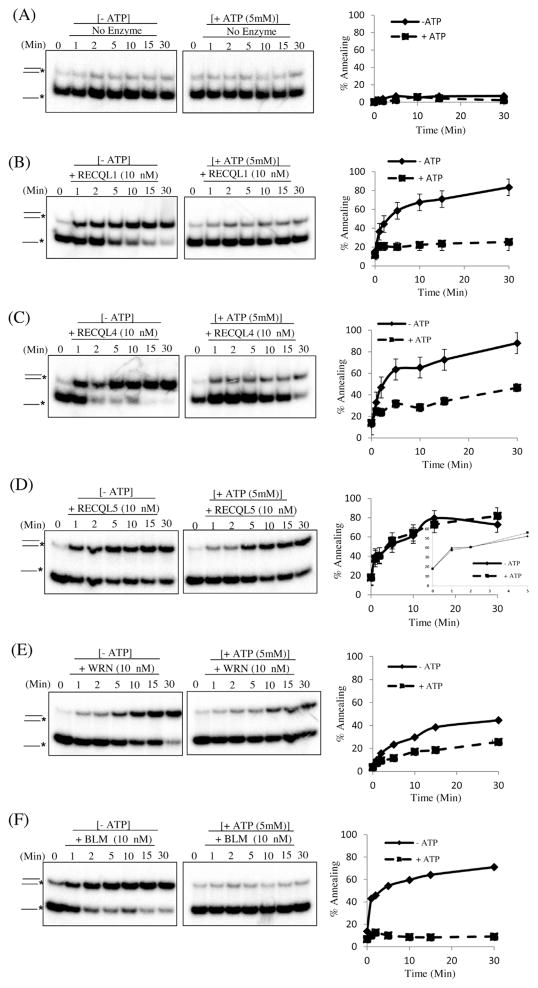
Recently, a number of studies have demonstrated that human RecQs possess strong DNA double strand annealing activity [1,11,27,37]. To compare this activity amongst the human RecQs proteins, we tested each human protein using the same reaction conditions (buffer, time and temperature) and used two different substrates. The single stranded DNA substrates, T3 and B3 (80-mers) or T2 and B2 (22-mers), were incubated with 10 nM purified RecQ protein in the presence or absence of ATP. The results demonstrate that all five human RecQs possess DNA strand annealing activity on both substrates (Figs. 1 and 2). There was no product formation in the absence of the protein during the time period of the reaction (Fig. 1A), indicating that the RecQ helicases promote DNA strand annealing. We analyzed the enzyme kinetics of each RecQ. We first evaluated the DNA strand annealing activity of RECQL1, RECQL4, WRN, BLM and RECQL5 at different time intervals (0, 1, 2, 5, 10, 15 and 30 min) in the presence or absence of ATP using oligonucleotides T3 and B3, see methods for a full description and Table 1 for the sequences. The annealed products were separated from ssDNA on a native polyacrylamide gel (Fig. 1B–F).
Fig. 1.
RecQ proteins promote strand annealing of complementary 80-mer ssDNA. (A) Time course of ssDNA annealing reactions with the indicated proteins, in the presence or absence of ATP. b–f, Time course of ssDNA annealing in reactions containing 10 nM RECQL1, RECQL4, RECQL5, WRN or BLM, respectively, in the presence or absence of ATP. The T3 and B3 complementary oligonucleotides (0.5 nM), of which B3 was radiolabeled on its 5′ end, were incubated with the indicated concentrations of RecQs proteins for the indicated time at 37 °C. The products of the annealing reactions were analyzed by gel electrophoresis. The positions of the unannealed ssDNA and the annealed fully duplex or partial duplex products are indicated on the left. In (A–F), the percentage of ssDNA annealed was quantified using three independent and the data are presented graphically on the right of the corresponding gel. Error bars represent standard deviation of three independent experiments.
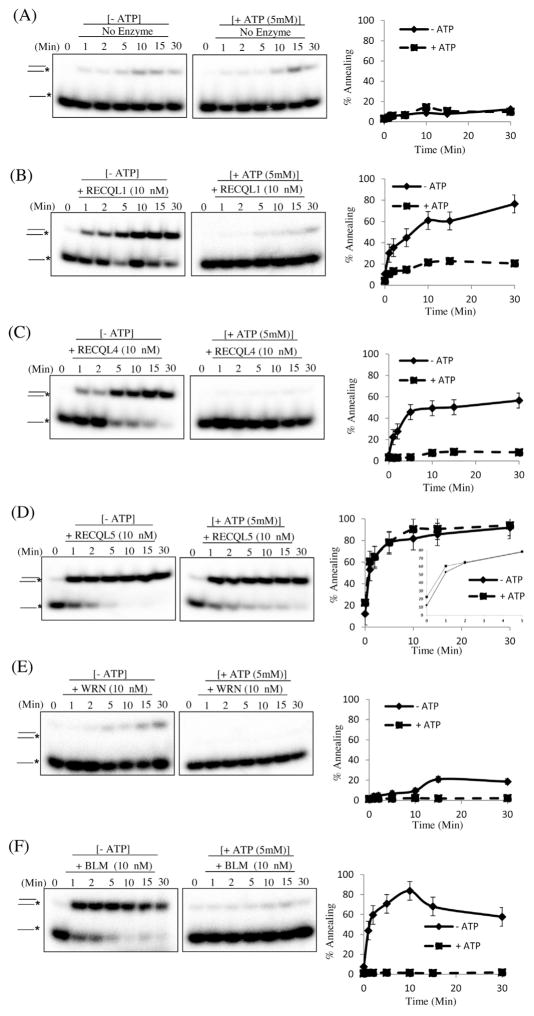
Fig. 2.
RecQ proteins promote annealing of 22-mer ssDNA. (A) Time course of ssDNA annealing in reactions containing the indicated protein, in the presence or absence of ATP. b–f, Time course of ssDNA annealing in reactions containing 10 nM RECQL1, RECQL4, RECQL5, WRN and BLM respectively, in the presence or absence of ATP. The T2 and B2 complementary oligonucleotides (0.5 nM), of which B2 was radiolabeled on its 5′ end, were incubated with the indicated concentrations of RecQs helicases for indicated time course at 37 °C. The products of annealing reactions were analyzed by gel electrophoresis. The positions of the unannealed ssDNA and the annealed full duplex or partial duplex products are indicated on the left. In (A–F), the percentage of ssDNA annealed was quantified using three independent experiments and the data are presented graphically on the right of the corresponding gel. Error bars represent standard deviation of three independent experiments.
Interestingly, RECQL1 and BLM had poor strand annealing in the presence of ATP while WRN and RECQL4 showed intermediate incapacitation after addition of ATP. Curiously and in contrast to the other proteins, RECQL5 possessed strong annealing activity even in the presence of ATP. The initial rate of strand annealing activity of RECQL5 is also decreased in the presence of ATP as compared to the absence of ATP (Fig. 1D). This shows that ATP might have a general inhibitory effect on the strand annealing activity of RecQ proteins.
3.2. Effect of oligonucleotide length on strand annealing by RecQ proteins
Next, we asked whether oligonucleotide length influenced the efficiency of each RecQ protein’s strand annealing by testing a shorter set of oligonucleotides, T2 and B2, see Table 1. We found that RECQL1 and BLM annealed complementary 22-mer oligonucleotides very efficiently under the same conditions and concentrations as for the 80-mer oligonucleotides (Fig. 1B and Fig. 2B). Previously, RECQL1 and BLM’s strand annealing activities were demonstrated on a forked heteroduplex [31]. Interestingly, RECQL4 and WRN performed strand annealing more efficiently on the longer substrates, (T3 & B3) than on the shorter substrate (T2 & B2) (Fig. 1 and Fig. 2, panels C and E). In contrast, RECQL5 performed strand annealing more efficiently with the shorter substrate than the longer substrate (Fig. 1D and Fig. 2D). However, we cannot exclude the possible effects of GC content in two different substrates. Nevertheless, this suggests that each of the RecQs has a different substrate preference, which may be important during DNA repair.
3.3. Effect of ATP and Mg2+ concentration on strand annealing by RecQ proteins
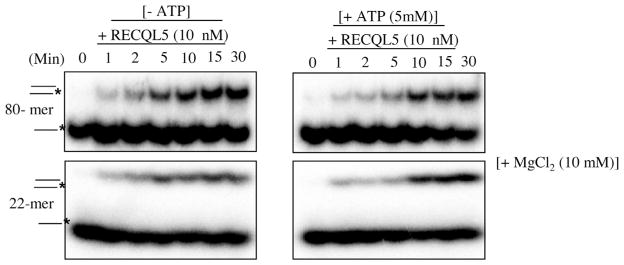
Previously it was shown that RECQL1 can anneal a fork duplex substrate in the presence of 2 mM ATP, but not in the presence of 5 mM ATP [31]. Therefore, we tested RECQL5’s relative strand annealing using different ATP concentrations from 0.5 mM to 4 mM rather than 5 mM. As shown in Supplementary Fig. S1, annealing by RECQL5 was not affected by ATP concentrations ranging from 0.5 mM to 4 mM. Similarly, it has been shown that RECQL1 helicase activity is sensitive to increasing ratio of Mg2+ to ATP [31], whereas, WRN helicase activity is not affected by increasing ratio of Mg2+ to ATP [38]. Thus, the strand annealing activity of RECQL5 was examined under conditions of greater Mg2+: ATP ratio up to 2. Strand annealing by RECQL5 was tested in the presence or absence of ATP in the presence of 10 mM MgCl2 using both long substrates (T3 & B3) and short substrates (T2 & B2). As shown in Fig. 3, RECLQ5 strand annealing activity was not affected by an increased Mg2+: ATP ratio. Similarly, there was change when we used 10 mM MgCl2 on the strand annealing activity of the other four RecQs (Supplementary Fig. 4). Thus RECQL5 can perform strand annealing in the presence or absence of ATP in the presence of MgCl2 (10 mM). RECQL5 is functionally distinct from the other human RecQ proteins, and this unique capability may enable it to play a specialized role during DNA repair.
Fig. 3.
RecQ proteins promote annealing of ssDNA under conditions of greater Mg2+:ATP ratio. (A) Time course of ssDNA annealing in reactions containing the indicated protein, in the presence or absence of ATP. Time course of ssDNA annealing in reactions containing 10 nM RECQL5, in the presence or absence of ATP under conditions of greater Mg2+:ATP ratio up to 2.The T3 and B3, T2 and B2 complementary oligonucleotides (0.5 nM), of which B3 and B2 were radiolabeled on its 5′ end, were incubated with the indicated concentrations of RecQ helicases for indicated time course at 37 °C. The products of annealing reactions were analyzed by gel electrophoresis.
3.4. Comparison of RecQ helicase and strand annealing activities
Next, we compared the helicase activity of each RecQ protein to directly compare their helicase to strand annealing activity (Supplementary Fig. S2). As shown in Supplementary Fig. S2A and B RECQL5 and RECQL4 have low helicase activities (based on concentration of RecQ proteins) whereas their strand annealing activity is strong (Figs. 1 and 2 and summarized in Table 2). Compared with RECQL5, RECQL4 shows low helicase activity even in the presence of ssDNA (data not shown). Similarly RECQL1, BLM and WRN show strong helicase activity (Supplementary Fig. S2C and E) and have a modest strand annealing activity compared to RECQL5 and RECQL4 proteins (Table 2). From this analysis it appears that it is not universally true that the strength of any given protein’s helicase and strand annealing activities are inversely correlated (with ATP present). While this statement may be true for the ‘strong’ helicases, like BLM and WRN, it does not hold for RECQL4.
Table 2.
Functional role of RecQ catalyzed strand annealing activity in the presence and absence of ATP.
| RecQ protein | Helicase activity | Strand Annealing Activity
|
|||
|---|---|---|---|---|---|
| 80-mer oligonucleotide
|
22-mer oligonucleotide
|
||||
| (−) ATP | (+) ATP | (−) ATP | (+) ATP | ||
| RECQL5 | + | ++++++ | +++ | +++ | +++ |
| RECQL4 | + | +++ | + | +++ | + |
| RECQL1 | ++ | ++ | + | ++ | + |
| BLM | +++ | +++ | + | +++ | + |
| WRN | +++ | +++ | + | +++ | + |
Note: Summary of strand annealing activity of the human RecQ proteins. DNA strand annealing was performed on two substrates (blunt end 80-mer and blunt end 22-mer), in the presence or absence of ATP. DNA helicase activity was performed using fork-1 duplex. (+) represents minimal activity, (++) represents good activity and (+++) represents the best activity.
3.5. Effects of known RecQ protein interacting partners on strand annealing
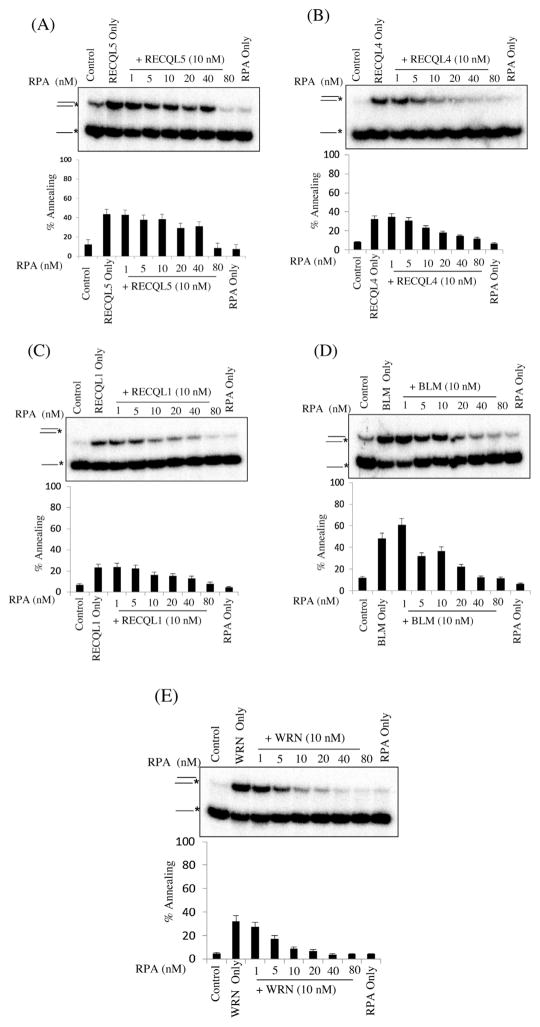
Previous studies have shown that the efficiency of RecQ strand annealing activity is modulated by several interacting partners that participate directly or indirectly in DSB repair or DNA metabolism (e.g. RPA, Ku, RAD51 and FEN1) [39–41]. We investigated whether known RecQ interacting partners modulates strand annealing activity similarly for each different RecQ protein in the absence of ATP. Previously, it was shown that RPA inhibits strand annealing by these enzymes [31] but usually a fork DNA duplex was used. Here we used single stranded 80-mer blunt end oligonucleotides, which better mimics the situation around double strand breaks in genome. As shown in Fig. 4A, the DNA strand annealing activity of RECQL5 was slightly inhibited by high concentrations of RPA (1:8). Similarly, we tested whether RECQL1, RECQL4, BLM and WRN were capable of strand annealing DNA in the presence of RPA [15,33,42,43]. Interestingly, as shown in Fig. 4B and E, RECQL4 and WRN catalyzed strand annealing was robustly inhibited in the presence of RPA, (1:1) ratio, whereas RECQL1 and BLM catalyzed strand annealing was inhibited in the presence of RPA at (1:4) ratio (Fig. 4C and D). RPA universally inhibits strand annealing mediated by the human RecQ proteins although the extent of inhibition varies among the proteins with RECQL5 being the least susceptible to this inhibition.
Fig. 4.
Effect of increasing concentrations of RPA on ssDNA annealing catalyzed by RecQ proteins. Panels A–E, strand annealing by the indicated RecQ protein (10 nM) as a function of increasing concentration of RPA, as indicated. RPA only lane contains maximum concentration. The T3 and B3 complementary oligonucleotides (0.5 nM), of which B3 was radiolabeled at its 5′ end, were incubated with the indicated concentrations of RecQ protein and increasing concentration of RPA for 10 min at 37 °C. Graph below shows quantification of the data. The positions of the unannealed ssDNA and the annealed full duplex or partial duplex products are indicated on the left. In A–E, the percentage of ssDNA annealed was quantified using three independent experiments and the data are presented graphically below of the corresponding dataset. Error bars represent standard deviation of three independent experiments.
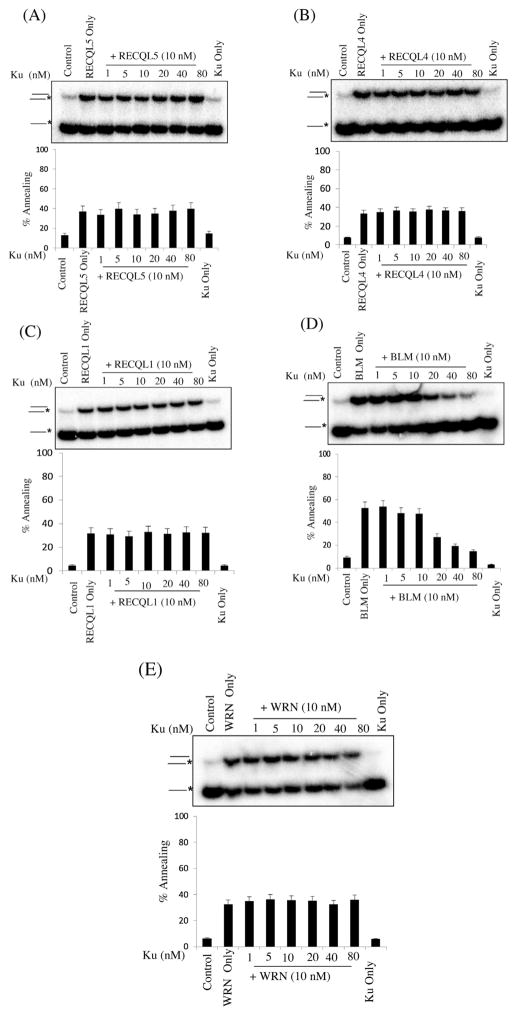
DSB are repaired by two major pathways: homologous recombination (HR) and classical non-homologous end joining (C-NHEJ) [29]. C-NHEJ relies on DNA-PK, Ku70/Ku80 and XRCC4/LIG IV, which generates small insertions or deletions when there is no complementary DNA strand available [28,29]. Ku binds to DSB ends and is one of the strongest interacting partners for WRN [44]. Besides, it has been shown that Ku interacts with RECQL1 and RECQL4 and modulates the double strand break repair by non-homologous end joining. Ku inhibits the helicase activity of RECQL4 but its effect on strand annealing of each RecQ has not been examined [45,46]. Thus, the impact of Ku on in vitro strand annealing of each RecQs was examined as shown in Fig. 5A–E, Ku strongly inhibits BLM’s strand annealing at high concentrations of Ku (1:4) (Fig. 4D) but it had no impact on any of the other human RecQs.
Fig. 5.
Effect of increasing concentrations of Ku on ssDNA annealing catalyzed by RecQ proteins. Panels A–E, strand annealing by the indicated RecQ protein (10 nM) as a function of increasing concentration of Ku, as indicated. Ku only lane contains maximum concentration. The T3 and B3 complementary oligonucleotides (0.5 nM), of which B3 was radiolabeled at its 5′ end, were incubated with the indicated concentrations of RecQ protein and increasing concentration of Ku for 10 min at 37 °C. Graph below shows quantification of the data. The positions of the unannealed ssDNA and the annealed full duplex or partial duplex products are indicated on the left. In A–E, the percentage of ssDNA annealed was quantified using three independent experiments and the data are presented graphically below of the corresponding dataset. Error bars represent standard deviation of three independent experiments.
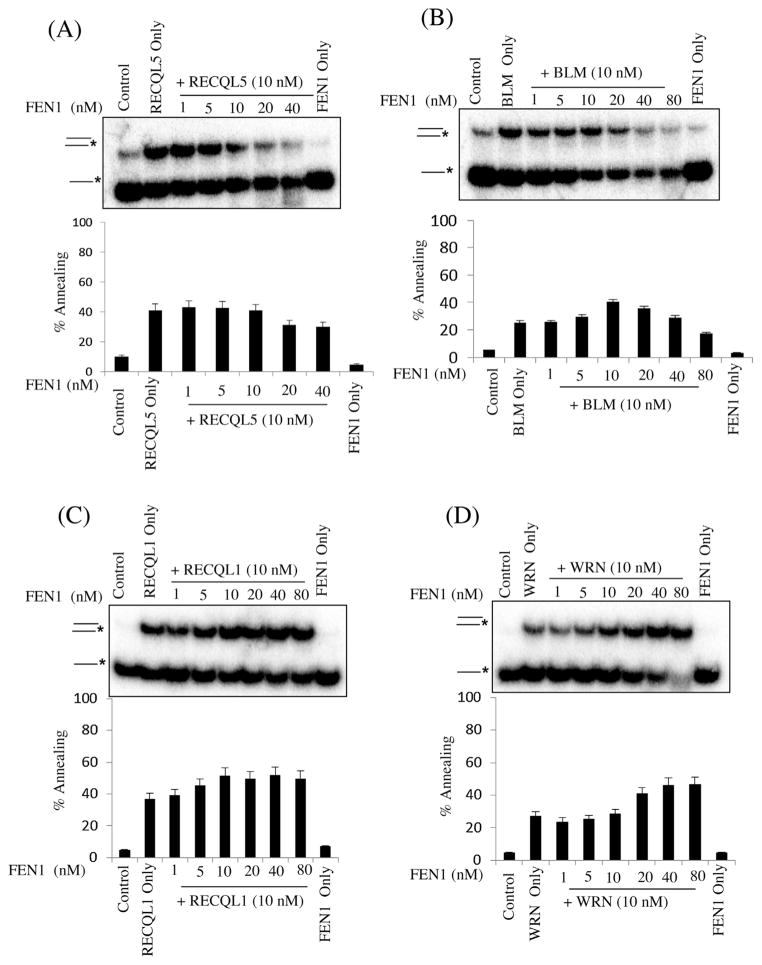
Next, we tested RecQ protein strand annealing activity in the presence of FEN1, as shown in Fig. 6A–D. FEN1 is known to associate with long-patch base excision repair (LP-BER) by cleaving DNA flap structures [47,48]. Recently, physical and functional interactions between FEN1 and WRN, BLM, RECQL4 and RECQL5 have been reported [21,49–51]. The mammalian RecQ proteins stimulated FEN1 cleavage activity. Therefore, we sought to analyze whether FEN1 protein could modulate RecQ proteins’ strand annealing activity. As shown in Fig. 6A–D, BLM and RECQL5 strand annealing activity was inhibited at high concentrations of FEN1 (1:4), whereas RECQL1 and WRN strand annealing activities were slightly stimulated at high concentrations of FEN1 (1:4). Thus, there does not appear to be a universal response on strand annealing due to the presence of FEN1 in the reactions.
Fig. 6.
Effect of increasing concentrations of FEN1 on ssDNA annealing catalyzed by RecQ proteins. Panel a–e, strand annealing by the indicated RecQ protein (10 nM) as a function of increasing concentration of FEN1, as indicated. FEN1 only lane contains maximum concentration. The T3 and B3 complementary oligonucleotides (0.5 nM), of which B3 was radiolabeled at its 5′ end, were incubated with the indicated concentrations of RecQ protein and increasing concentration of FEN1 for 10 min at 37 °C. Graph below shows quantification of the data. The positions of the unannealed ssDNA and the annealed full duplex or partial duplex products are indicated on the left. In A–E, the percentage of ssDNA annealed was quantified using three independent experiments and the data are presented graphically below of the corresponding dataset Error bars represent standard deviation of three independent experiments.
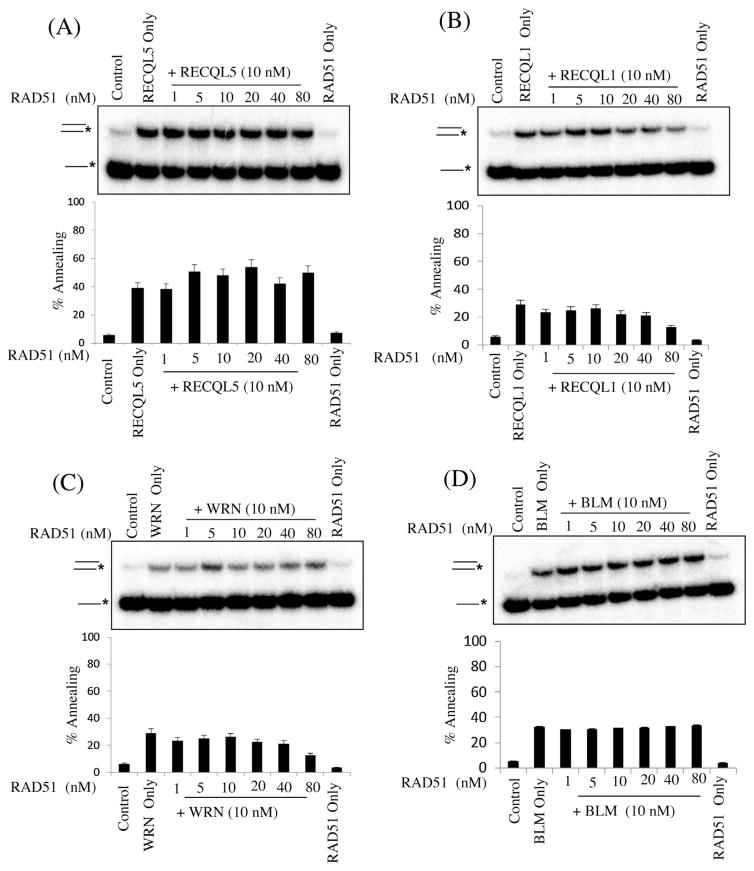
RAD51 is a recombinase that nucleates onto 3′ single DNA strand ends to initiate invasion onto a homologous template, usually provided by the complementary sister chromatid during replication maintaining replication forks and repairs DSBs [41,52,53]. RECQL5 can disrupt RAD51 nucleoprotein filaments, promoting noncrossover products during DNA double-strand break-induced HR and counteracts the inhibitory effect of RAD51 on RAD52-mediated DNA annealing in vitro and in vivo [41]. We performed strand annealing using RECQL5, RECQL1, BLM and WRN in the presence of RAD51 [54–56]. Interestingly, RAD51 had no effect on human RecQ protein catalyzed strand annealing activities (Fig. 7A–D).
Fig. 7.
Effect of increasing concentrations of RAD51 on ssDNA annealing catalyzed by RecQ proteins. Panels A–E, strand annealing by the indicated RecQ protein (10 nM) as a function of increasing concentration of RAD51, as indicated. Rad51 only lane contains maximum concentration. The T3 and B3 complementary oligonucleotides (0.5 nM), of which B3 was radiolabeled at its 5′ end, were incubated with the indicated concentrations of RecQ protein and increasing concentration of RAD51 for 10 min at 37 °C. Graph below shows quantification of the data. The positions of the unannealed ssDNA and the annealed full duplex or partial duplex products are indicated on the left. In a–e, the percentage of ssDNA annealed was quantified using three independent experiments and the data are presented graphically below of the corresponding dataset error bars represent standard deviation of three independent experiments.
A summary of each human RecQ protein’s strand annealing activity in the presence of key interacting partners is shown in Table 3, with + and − signs representing the relative stimulation and inhibition of strand annealing activity. These functional protein interactions may have implications on DSB repair.
Table 3.
Impact of interacting proteins on RecQ catalyzed strand annealing activity.
| Interacting protein | Strand annealing activity
|
|||
|---|---|---|---|---|
| RECQL5 | RECQL1 | BLM | WRN | |
| RPA | – | - - - | - - | - - - |
| Ku | NA | NA | - - | NA |
| FEN1 | - - | NA | - - | + |
| RAD51 | NA | – | NA | NA |
Note: Summary of DNA strand annealing activity of human RecQ proteins in the presence of RecQs interacting proteins. DNA strand annealing was performed on 80-mer substrates in the absence of ATP. (+) represents the minimal stimulation, (–) represents the minimal inhibition, (- -) represent greater inhibition, (- - -) represents the greatest inhibition and NA represent as not affected.
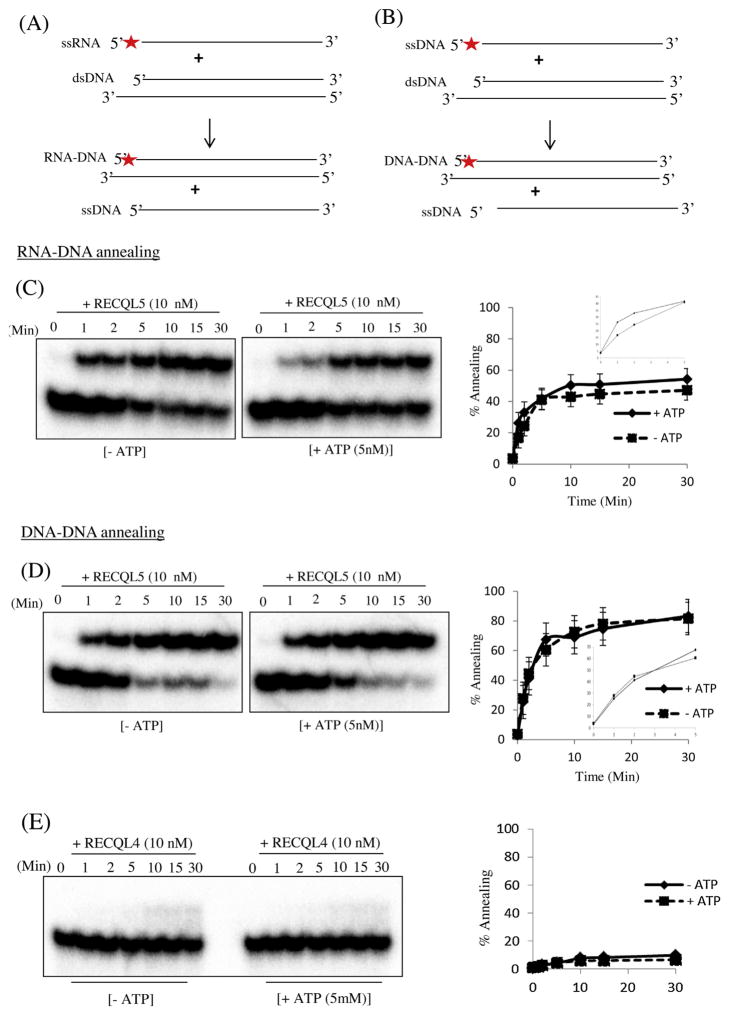
3.6. RECQL5 promotes DNA–DNA strand exchange and RNA–DNA strand exchange
RECQL5 is associated with RNA polymerase II (RNAPII) and is required as an elongation factor preserving the genome integrity during transcription [22,57]. RECQL5 promotes repair of DNA double-strand breaks via synthesis-dependent strand annealing pathway by disrupting RAD51 nucleoprotein filaments which promotes formation of non-crossover products during DNA double-strand break-induced HR and counteracts the inhibitory effect of RAD51 on RAD52-mediated DNA annealing [41]. Previous studies showed that E. coli RecA (homologous protein of human RAD51) promotes annealing between duplex DNA and single-strand RNA in vitro [58–60]. Therefore, we tested whether RECQL5 could promote a similar reaction. Indeed, RECQL5 promotes annealing of ssRNA to DNA as well as ssDNA to DNA (Fig. 8A–D). RECQL5 catalyzed the reaction between ssRNA and dsDNA at nearly half the rate of that between ssDNA and dsDNA (Fig. 8A–D). In the presence of ATP and ssRNA, the extent of strand exchange catalyzed by RECQL5 between ssRNA and dsDNA was about 40%, which is less than that between ssDNA and the dsDNA duplex. In contrast, the extent of the reaction catalyzed by RECQL5 using ssRNA and dsDNA was affected during the initial time of the reaction (0–5 min) in the presence ATP whereas, the extent of the reaction catalyzed by RECQL5 using ssDNA and dsDNA was not affected by the presence or absence of ATP. Thus, RECQL5 promotes strand exchange between ssDNA and dsDNA with higher efficiency than between ssRNA and dsDNA.
Fig. 8.
RECQL5 promotes strand exchange between RNA and DNA. (A) Experimental scheme of RECQL5 promoted strand exchange between RNA and DNA in vitro. (B) The strand exchange reactions were promoted by RECQL5 (10 nM) in the presence or absence of ATP (5 nM). dsDNA containing a protruding ssDNA tail (T4 and B4) was incubated with protein dilution buffer and then RECQL5 was added to the mixture. To initiate the annealing reactions, 0.5 nM 32P-end labelled ssRNA (T6) was added. The reactions were carried out for the indicated periods of time course, and the products of the annealing reactions were analyzed by electrophoresis. The percentage of ssRNA annealed was quantified using three independent experiments and the data are presented graphically below of the corresponding dataset. (C) Experimental scheme of RECQL5 promoted strand exchange between ssDNA and dsDNA in vitro. dsDNA containing a protruding ssDNA tail (T4 and B4) was incubated with protein dilution buffer and then RECQL5 was added to the mixture. To initiate the annealing reactions, 0.5 nM 32P-end labelled ssDNA (T5) were added. The percentage of ssDNA annealed was quantified using three independent experiments and the data are presented graphically below of the corresponding dataset. Error bars represent standard deviation of three independent experiments.
We asked whether the other human RecQ proteins (RECQL1, WRN, BLM and RECQL4,) could also catalyze the strand exchange of RNA to DNA. Strand exchange assays were performed with each RecQ protein at different time intervals (0, 1, 2, 5, 10, 15 and 30 min). The annealed products were separated from ssDNA on native polyacrylamide gels (Supplementary Fig. S3A and B). Interestingly, none of the other RecQ proteins were able to catalyze strand exchange of RNA to DNA as did RECQL5.
4. Discussion
RecQs helicases are involved in replication, DNA repair, telomere maintenance and transcription [27,37,61–63]. They can act on various substrates including fork and bubble structures, D-loops, Holliday junctions and G-quadruplex DNA [16,31]. They possess 3′–5′ helicase activity as well as ATP-independent single strand annealing activities [16,31,64,65]. Here, we compared the strand annealing activities of the five human RecQs proteins under the same buffer conditions and at the same protein concentrations. Interestingly, amongst all human RecQs, RECQL5 possesses relatively strong strand annealing activity in the presence or absence of ATP compared to the other RecQ proteins (Figs. 1 and 2). Importantly, we show that the strand annealing activity of RECQL5’s is not affected by ATP at the later time points whereas the annealing activities of all the other RecQ’s were inhibited by the presence of ATP throughout the course of the reactions. This shows that RECQL5 is unique and that its strand annealing mechanism is different from that of the other human proteins.
Previously it has been suggested that nucleotide-induced conformational change in the RECQL1 protein alters the DNA unwinding mode to strand annealing [66]. Partial proteolysis studies demonstrated that ATP binding induces a conformational change in RECQL1 protein which serves as a molecular switch allowing it to change from a strand-annealing to a DNA-unwinding mode [67]. A form identified as monomers or dimers is responsible for DNA unwinding activity, whereas higher-order oligomers (hexamers or pentamers) possess strand-annealing activity (23). Moreover, ATP binding is the key that controls the equilibrium between these two assembly states, favoring the smaller form [67,68]. Whereas all the other RecQs have been reported to exist in higher order complexes, this does not seem to be the case for RECQL5, and this may explain why RECQL5 is impervious to ATP. Since RECQL5 is reported to be a monomer, the binding of ATP, DNA and RECQL5 might not affect the structure of RECQL5 protein and alter its strand annealing activity [2,66,67]. It will be interesting to further characterize the RECQL5 crystal structure in complex with DNA or RNA molecules which could elucidate RECQL5’s mechanisms of action.
For proper function of the RecQs helicases, their enzymatic action need to be tightly controlled because unnecessary unwinding of DNA can cause genome instability. Thus, the enzymatic action of RecQs helicase’s can be controlled either by their interacting partners or post-translational modifications (PTMs) [1]. RECQL5 physically and functionally interacts with WRN both in vivo and in vitro and co-operates with it on synthetic stalled replication fork-like structures, and stimulates its helicase activity on DNA fork duplexes [69]. Similarly, RECQL4 interacts with BLM both in vitro and in vivo and specifically stimulates BLM helicase activity on DNA fork substrates in vitro [16]. RPA is known to inhibit RECQL1 and RECQL5 mediated strand annealing of fork duplexes [31,65] while it stimulates the helicase activity of RECQL4 and BLM, respectively [33,70]. Here, we compared the strand annealing activity of RecQs helicases in the presence of various interacting proteins (Figs. 4–7). RPA universally inhibits the strand annealing activity of human RecQ proteins. However, notably, RECQL5’s strand annealing activity was only slightly inhibited at high RPA concentrations (1:8 protein concentration ratios) whereas all other human RecQ proteins were inhibited at low concentrations of RPA (1:1) (Fig. 4A–E). In the presence of Ku, BLM strand annealing activity was inhibited at high Ku concentrations (1:8) but there was no effect on the other four RecQ protein’s strand annealing activity (Fig. 5A–E). Similarly, the strand annealing activity of the human RecQ’s were not affected by FEN1 or RAD51, respectively (Figs. 6 and 7). This shows that each of the RecQs interacting proteins (direct or indirectly) have different functional interaction.
Efficient DNA repair of all DNA lesions is essential to maintain genome integrity and cell viability. Deficiencies and mutations in proteins involved in HR and NHEJ pathways are associated with human diseases and aging. Homologous recombination, the exchange of DNA between homologous DNA molecules, is a central process for repair which involves all five RecQ proteins at different levels in the repair pathway. Inappropriate DNA rearrangements can lead to genomic instability which needs to be avoided as it can be deleterious. Thus cells need to balance DNA repair, an important feature required to maintain the genome stability [61]. Of the five RecQ proteins, RECQL5 and RECQL4 show very weak helicase activity relative to RECQL1, BLM and WRN. In contrast, RECQL5 and RECQL4 possess relatively strong strand annealing activity (both with long substrate and short substrate) than that of RECQL1, BLM and WRN (under our experimental conditions). (Table 2). We conclude that annealing activity is not just the inverse to helicase activity is of importance and future work should be more focused on identifying the in vivo function of strand annealing and its role in DNA repair related pathways. Aside from its DNA repair implication, an understanding of the strand annealing activity of RecQ proteins is essential for the DNA replication. The formation of duplex DNA via strand annealing activity is a fundamental biological process that is required for genome stability and for genome evolution.
RECQL5 is associated with RNA polymerase II (RNAPII) and is required as an elongation factor [57]. RECQL5-deficient cells exhibit elevated frequencies of spontaneous DNA double-strand breaks and homologous recombination and are prone to gross chromosomal rearrangements in response to a stalled replication fork progression [25]. We tested strand exchange by RECQL5 on DNA–DNA as well as on RNA–DNA substrates. RECQL5 annealed ssDNA to complementary dsDNA more efficiently than ssRNA to dsDNA substrates (Fig. 8A–C). Thus from our in vitro data, it appears that RECQL5 uniquely can promote RNA–DNA annealing. Perhaps RECQL5 utilizes this mechanism to slow down RNA polymerase transcription. Interestingly, we found that amongst all five human RecQ helicases only RECQL5 has RNA–DNA exchange activity (Supplementary Fig. S3), which further shows that RECQL5 is unique.
RECQL5 is known to unwind the lagging strand arm and to promote strand exchange on hRPA coated forked duplex structure [25,65]. Similarly, RECQL5 also regulates HR by binding to RAD51 and inhibiting the RAD51 mediated D-loop formation [25]. RECQL5 displaces RAD51 from single-stand DNA in a reaction that requires ATP hydrolysis and RPA [25]. We show here that RECQL5 can perform both DNA unwinding and annealing in the presence of ATP. The precise coordination of these two activities of RECQL5 is central to its role in various DNA repair pathways. However, the biological significance of strand annealing by RECQL5 and its role in vivo still require investigation. It would be interesting to further characterize these dueling properties of RECQL5 in a cell based assay if one was available. RECQL5 has to perform unwinding and annealing depending on the specific substrate, cellular conditions, and the DNA DSB repair pathway. ATP hydrolysis is required for unwinding DNA substrates whereas ATP inhibits the strand annealing activity of human RecQs helicase with the exception of RECQL5. It is possible that RECQL5’s strand annealing activity is required when the other four human RecQ proteins cannot perform annealing.
Interestingly, in a yeast system a novel mechanism of homologous recombination repair in which transcript RNA is used as a template for DSB repair and using the strand annealing properties of a helicase protein has been reported [22]. The implications are that RNA transcripts can template DNA damage repair, in cells that do not divide or have more stable RNA–DNA heteroduplex, such as in cells with defective RNASE H2 in patients with Aicardi Goutie’res syndrome [22]. It is tempting to speculate that RECQL5 might remodel DNA–RNA structures or nucleoprotein complexes after collusion of the transcription and replication machineries. Thus, it will be interesting to explore further whether RECQL5 has a role in RNA-driven DNA recombination and repair during DSB in human cells.
Supplementary Material
Highlights.
The RecQ helicases all have strand annealing activities, very important in genome maintenance.
We have compared the helicase and strand annealing properties of all 5 human RecQ helicases.
RECQ5 has unique annealing properties including RNA:DNA annealing.
Acknowledgments
This work was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging. We would like to thank individuals that contributed to the various protein purifications: Dr. Takashi Tadokoro and Dr. Tomasz Kulikowicz and Christopher Dun.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.dnarep.2015.11.005.
References
- 1.Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Ann Rev Biochem. 2014;83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett RJ, Keck JL. Structure and function of RecQ DNA helicases. Crit Rev Biochem Mol Biol. 2004;39:79–97. doi: 10.1080/10409230490460756. [DOI] [PubMed] [Google Scholar]
- 3.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 4.Brosh RM., Jr DNA helicases involved in DNA repair and their roles in cancer. Nat Rev Cancer. 2013;13:542–558. doi: 10.1038/nrc3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobb JA, Bjergbaek L. RecQ helicases: lessons from model organisms. Nucleic Acids Res. 2006;34:4106–4114. doi: 10.1093/nar/gkl557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mackintosh SG, Raney KD. DNA unwinding and protein displacement by superfamily 1 and superfamily 2 helicases. Nucleic Acids Res. 2006;34:4154–4159. doi: 10.1093/nar/gkl501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Ann Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 9.Brosh RM, Jr, Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007;35:7527–7544. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, Hickson ID, Vindigni A. The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem. 2008;283:17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- 11.Killoran MP, Keck JL. Sit down relax and unwind: structural insights into RecQ helicase mechanisms. Nucleic Acids Res. 2006;34:4098–4105. doi: 10.1093/nar/gkl538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachrati CZ, Hickson ID. RecQ Helicases: suppressors of tumorigenesis and premature aging. Biochem J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khakhar R. RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol. 2003;13:493–501. doi: 10.1016/s0962-8924(03)00171-5. [DOI] [PubMed] [Google Scholar]
- 14.Monnat RJ., Jr Human RECQ helicases: roles in DNA metabolism, mutagenesis and cancer biology. Semi Cancer Biol. 2010;20:329–339. doi: 10.1016/j.semcancer.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheok CF, Wu L, Garcia PL, Janscak P. The Bloom’s syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005;33:3932–3941. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh DK, Popuri V, Kulikowicz T, Shevelev I, Ghosh AK, Ramamoorthy M, Rossi ML, Janscak P, Croteau DL, Bohr VA. The human RecQ helicases BLM and RECQL4 cooperate to preserve genome stability. Nucleic Acids Res. 2012;40:6632–6648. doi: 10.1093/nar/gks349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tadokoro T, Ramamoorthy M, Popuri V, May A, Tian J, Sykora P, Rybanska I, Wilson DM, 3rd, Croteau DL, Bohr VA. Human RECQL5 participates in the removal of endogenous DNA damage. Mol Biol Cell. 2012;23:4273–4285. doi: 10.1091/mbc.E12-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin Y, Wang W, Monnat RJ, Jr, Falaschi A, Vindigni A. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol Cell Biol. 2010;30:1382–1396. doi: 10.1128/MCB.01290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popuri V, Hsu J, Khadka P, Horvath K, Liu Y, Croteau DL, Bohr VA. Human RECQL1 participates in telomere maintenance. Nucleic Acids Res. 2014;42:5671–5688. doi: 10.1093/nar/gku200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popuri V, Tadokoro T, Croteau DL, Bohr VA. Human RECQL5: guarding the crossroads of DNA replication and transcription and providing backup capability. Crit Rev Biochem Mol Biol. 2013;48:289–299. doi: 10.3109/10409238.2013.792770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Speina E, Dawut L, Hedayati M, Wang Z, May A, Schwendener S, Janscak P, Croteau DL, Bohr VA. Human RECQL5beta stimulates flap endonuclease 1. Nucleic Acids Res. 2010;38:2904–2916. doi: 10.1093/nar/gkp1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keskin H, Shen Y, Huang F, Patel M, Yang T, Ashley K, Mazin AV, Storici F. Transcript-RNA-templated DNA recombination and repair. Nature. 2014;515:436–439. doi: 10.1038/nature13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aygun O, Svejstrup JQ. RECQL5 helicase: connections to DNA recombination and RNA polymerase II transcription. DNA Rep. 2010;9:345–353. doi: 10.1016/j.dnarep.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Aygun O, Xu X, Liu Y, Takahashi H, Kong SE, Conaway RC, Conaway JW, Svejstrup JQ. Direct inhibition of RNA polymerase II transcription by RECQL5. J Biol Chem. 2009;284:23197–23203. doi: 10.1074/jbc.M109.015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, Jasin M, Vogel H, Sung P, Luo G. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Y, Lu X, Luo G. Effect of Recql5 deficiency on the intestinal tumor susceptibility of apcminmice. World J Gastroenterol. 2010;16:1482. doi: 10.3748/wjg.v16.i12.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y. Unwinding and rewinding: double faces of helicase? J Nucleic Acids. 2012;2012:140601. doi: 10.1155/2012/140601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C, Srihari S, Cao KA, Chenevix-Trench G, Simpson PT, Ragan MA, Khanna KK. A fine-scale dissection of the DNA double-strand break repair machinery and its implications for breast cancer therapy. Nucleic Acids Res. 2014;42:6106–6127. doi: 10.1093/nar/gku284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Ann Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 30.Janscak P, Garcia PL, Hamburger F, Makuta Y, Shiraishi K, Imai Y, Ikeda H, Bickle TA. Characterization and mutational analysis of the RecQ core of the bloom syndrome protein. J Mol Biol. 2003;330:29–42. doi: 10.1016/s0022-2836(03)00534-5. [DOI] [PubMed] [Google Scholar]
- 31.Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, Muzzolini L, Vindigni A, Brosh RM., Jr Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 32.Orren DK, Brosh RM, Jr, Nehlin OJ, Machwe A, Gray DM, Bohr VA. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossi ML, Ghosh AK, Kulikowicz T, Croteau DL, Bohr VA. Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding. DNA Rep. 2010;9:796–804. doi: 10.1016/j.dnarep.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.B-IL, MDW The RAD2 domain of human exonuclease 1 exhibits 5′–3′ exonuclease and flap structure-specific endonuclease activities. J Biol Chem. 1999;274:37763–37769. doi: 10.1074/jbc.274.53.37763. [DOI] [PubMed] [Google Scholar]
- 35.Binz SK, Dickson AM, Haring SJ, Wold MS. Functional assays for replication Protein A (RPA) 2006;409:11–38. doi: 10.1016/S0076-6879(05)09002-6. [DOI] [PubMed] [Google Scholar]
- 36.Sigurdsson S, Van Komen S, Petukhova G, Sung P. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J Biol Chem. 2002;277:42790–42794. doi: 10.1074/jbc.M208004200. [DOI] [PubMed] [Google Scholar]
- 37.Wu L, Hickson ID. DNA helicases required for homologous recombination and repair of damaged replication forks. Ann Rev Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- 38.Choudhary S, Sommers JA, Brosh RM., Jr Biochemical and kinetic characterization of the DNA helicase and exonuclease activities of werner syndrome protein. J Biol Chem. 2004;279:34603–34613. doi: 10.1074/jbc.M401901200. [DOI] [PubMed] [Google Scholar]
- 39.Deng SK, Gibb B, de Almeida MJ, Greene EC, Symington LS. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:405–412. doi: 10.1038/nsmb.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finn KJ, Li JJ. Single-stranded annealing induced by re-initiation of replication origins provides a novel and efficient mechanism for generating copy number expansion via non-allelic homologous recombination. PLoS Genet. 2013;9:e1003192. doi: 10.1371/journal.pgen.1003192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paliwal S, Kanagaraj R, Sturzenegger A, Burdova K, Janscak P. Human RECQ5 helicase promotes repair of DNA double-strand breaks by synthesis-dependent strand annealing. Nucleic Acids Res. 2014;42:2380–2390. doi: 10.1093/nar/gkt1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Machwe A, Lozada EM, Xiao L, Orren DK. Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins. BMC Mol Biol. 2006;7:1. doi: 10.1186/1471-2199-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meltem TK, Beck MG, Lee JW, Piotrowski J, Bohr VA. Intrinsic ssDNA annealing activity in the C-terminal region of WRN. Biochemistry. 2008;47:10247–10254. doi: 10.1021/bi800807n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B, Comai L. Functional interaction between Ku and the werner syndrome protein in DNA end processing. J Biol Chem. 2000;275:28349–28352. doi: 10.1074/jbc.C000289200. [DOI] [PubMed] [Google Scholar]
- 45.Parvathaneni S, Stortchevoi A, Sommers JA, Brosh RM, Jr, Sharma S. Human RECQ1 interacts with Ku70/80 and modulates DNA end-joining of double-strand breaks. PloS One. 2013;8:e62481. doi: 10.1371/journal.pone.0062481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shamanna RA, Singh DK, Lu H, Mirey G, Keijzers G, Salles B, Croteau DL, Bohr VA. RECQ helicase RECQL4 participates in non-homologous end joining and interacts with the Ku complex. Carcinogenesis. 2014;35:2415–2424. doi: 10.1093/carcin/bgu137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prasad R, Lavrik OI, Kim SJ, Kedar P, Yang XP, Vande Berg BJ, Wilson SH. DNA polymerase beta -mediated long patch base excision repair. Poly(ADP-ribose) polymerase-1 stimulates strand displacement DNA synthesis. J Biol Chem. 2001;276:32411–32414. doi: 10.1074/jbc.C100292200. [DOI] [PubMed] [Google Scholar]
- 48.Klungland Aa, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma S, Sommers JA, Wu L, Bohr VA, Hickson ID, Brosh RM., Jr Stimulation of flap endonuclease-1 by the bloom’s syndrome protein. J Biol Chem. 2004;279:9847–9856. doi: 10.1074/jbc.M309898200. [DOI] [PubMed] [Google Scholar]
- 50.Brosh RM, Jr, von Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GLa, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schurman SH, Hedayati M, Wang Z, Singh DK, Speina E, Zhang Y, Becker K, Macris M, Sung P, Wilson DM, 3rd, Croteau DL, Bohr VA. Direct and indirect roles of RECQL4 in modulating base excision repair capacity. Hum Mol Genet. 2009;18:3470–3483. doi: 10.1093/hmg/ddp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakano A, Masuda K, Hiromoto T, Takahashi K, Matsumoto Y, Habib AG, Darwish AG, Yukawa M, Tsuchiya E, Ueno M. Rad51-dependent aberrant chromosome structures at telomeres and ribosomal DNA activate the spindle assembly checkpoint. Mol Cell Biol. 2014;34:1389–1397. doi: 10.1128/MCB.01704-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hosono Y, Abe T, Ishiai M, Islam MN, Arakawa H, Wang W, Takeda S, Ishii Y, Takata M, Seki M, Enomoto T. Tumor suppressor RecQL5 controls recombination induced by DNA crosslinking agents. Biochim Biophys Acta. 2014;1843:1002–1012. doi: 10.1016/j.bbamcr.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Budke B, Chan YL, Bishop DK, Connell PP. Real-time solution measurement of RAD51- and RecA-mediated strand assimilation without background annealing. Nucleic Acids Res. 2013;41:e130. doi: 10.1093/nar/gkt362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neal Sugawara GIAJEH. DNA length dependecnce of the single-strand anneling pathway and the role of saccharomyces cerevisiae RAD59 in double-strand break repair. Mol Cell Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Serra H, Da Ines O, Degroote F, Gallego ME, White CI. Roles of XRCC2, RAD51B and RAD51D in RAD51-independent SSA recombination. PLoS Genet. 2013;9:e1003971. doi: 10.1371/journal.pgen.1003971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Izumikawa K, Yanagida M, Hayano T, Tachikawa H, Komatsu W, Shimamoto A, Futami K, Furuichi Y, Shinkawa T, Yamauchi Y, Isobe T, Takahashi N. Association of human DNA helicase RecQ5beta with RNA polymerase II and its possible role in transcription. Biochem J. 2008;413:505–516. doi: 10.1042/BJ20071392. [DOI] [PubMed] [Google Scholar]
- 58.Kasahara M, Clikeman AJ, Bates BD, Kogoma T. RecA protein-dependent R Loop formation in vitro, Genes. Development. 1999;36:360–365. [PMC free article] [PubMed] [Google Scholar]
- 59.WL, GKS, KD The homologous recombination machinery modulates the formation of RNA–DNA hybrids and associated chromosome instability. Elife. 2013:e00505. doi: 10.7554/eLife.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaitsev NE, Smith KC. A novel pairing process promoted by Escherichia coli RecA protein: in verse DNA and RNA strand exchange. Genes Dev. 2000:740–749. [PMC free article] [PubMed] [Google Scholar]
- 61.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 62.Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Ann Rev Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 64.Keller H, Kiosze K, Sachsenweger J, Haumann S, Ohlenschlager O, Nuutinen T, Syvaoja JE, Gorlach M, Grosse F, Pospiech H. The intrinsically disordered amino-terminal region of human RecQL4: multiple DNA-binding domains confer annealing, strand exchange and G4 DNA binding. Nucleic Acids Res. 2014;42:12614–12627. doi: 10.1093/nar/gku993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garcia LP, Liu Y, Jiricny J, West CSa, Janscak P. Human RECQ5B, a protein with DNA helicase and strand annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lucic B, Zhang Y, King O, Mendoza-Maldonado R, Berti M, Niesen FH, Burgess-Brown NA, Pike AC, Cooper CD, Gileadi O, Vindigni A. A prominent beta-hairpin structure in the winged-helix domain of RECQ1 is required for DNA unwinding and oligomer formation. Nucleic Acids Res. 2011;39:1703–1717. doi: 10.1093/nar/gkq1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pike AC, Gomathinayaram S, Swuec P, Berti M, Zhang Y, Schnecke C, Marino F, Delft vF, Renault L, Costa A, Gileadi O, Vindigni A. Human RECQ1 helicase driven DNA unwinding, annealing, and branch migration Insights from DNA complex structures. PNAS. 2015;112:4286–4291. doi: 10.1073/pnas.1417594112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharma S, Doherty KM, Brosh RM., Jr Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Popuri V, Huang J, Ramamoorthy M, Tadokoro T, Croteau DL, Bohr VA. RECQL5 plays co-operative and complementary roles with WRN syndrome helicase. Nucleic Acids Res. 2013;41:881–899. doi: 10.1093/nar/gks1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.