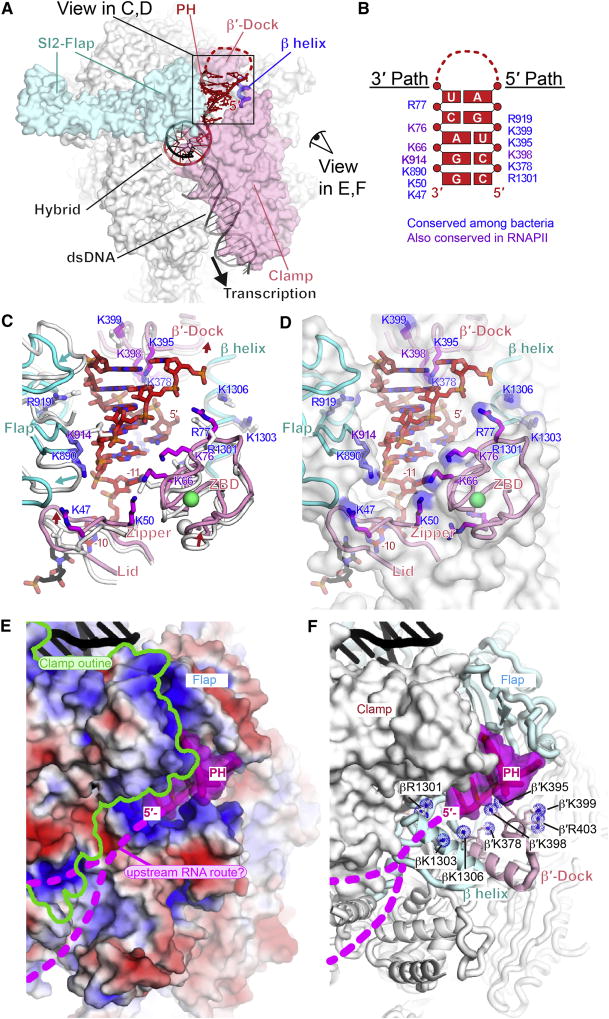
Figure 6. his PH-RNAP exit channel interaction.
(A) Interactions of PH in RNA exit channel. Flap, β′-dock, β helix, and clamp cradle the PH. Views in panels C-F are indicated.
(B) Map of side chains that form 3′ and 5′ paths complementary to PH. Purple, conserved among all RNAPs; blue, conserved only among bacterial RNAPs (Table S4).
(C) Movements in the RNA exit channel upon PH formation and key PH-interacting residues. The positions of exit channel modules (flap, lid, zipper, ZBD, β′-dock and β helix) in the hisPEC are shown as colored cartoons (cyan, β; pink, β′) and their corresponding positions in the EC are shown as white cartoons. The last bp of the RNA-DNA hybrid and the spacer −11 nt between the hybrid and the PH are shown. Positively charged residues that interact with the PH are shown as sticks (blue, β subunit; magenta, β′ subunit) with labels color-coded as in panel B.
(D) Same as panel C, except that the EC is not shown and the exit channel is shown as a semi-transparent, white surface with blue patches of positive charge.
(E) View from outside RNA exit channel and underside of the clamp with the surface of RNAP colored by electrostatic charge (−5 red to +5 blue). Upstream DNA is black. The PH is shown as a cartoon with a semi-transparent magenta surface and possible routes of ssRNA upstream from the PH are shown as dotted magenta lines (called “groove 1” in Cramer et al., 2001).
(F) Same view as panel E, but with a solid white RNAP surface except for the flap, β′-dock, and β-helix shown as cartoons and positive charges shown as blue dotted spheres.
Also see Table S4 and Figure S7.