Abstract
The molecular mechanism underlying the initiation of somatic cell reprogramming into induced pluripotent stem cells (iPSCs) has not been well described. Thus, we generated single-cell-derived clones by using a combination of drug-inducible vectors encoding transcription factors (Oct4, Sox2, Klf4 and Myc) and a single-cell expansion strategy. This system achieved a high reprogramming efficiency after metabolic and epigenetic remodeling. Functional analyses of the cloned cells revealed that extracellular signal-regulated kinase (ERK) signaling was downregulated at an early stage of reprogramming and that its inhibition was a driving force for iPSC formation. Among the reprogramming factors, Myc predominantly induced ERK suppression. ERK inhibition upregulated the conversion of somatic cells into iPSCs through concomitant suppression of serum response factor (SRF). Conversely, SRF activation suppressed the reprogramming induced by ERK inhibition and negatively regulated embryonic pluripotency by inducing differentiation via upregulation of immediate early genes, such as c-Jun, c-Fos and EGR1. These data reveal that suppression of the ERK-SRF axis is an initial molecular event that facilitates iPSC formation and may be a useful surrogate marker for cellular reprogramming.
Introduction
The reprogramming mechanisms by which somatic cells acquire embryonic stem cell (ESC) properties, such as self-renewal and pluripotency, have been intensively studied at the molecular level.1, 2, 3, 4, 5 Among these, elucidation of the molecular mechanisms underlying the early steps of reprogramming may provide insight into this phenomenon, which is the reverse of the steps that occur during development, and help to identify other means by which to acquire self-renewal and pluripotency. However, the molecular characteristics of this phase are poorly defined in comparison with those of the middle/late stages, which drive epigenetic changes in somatic cells.3 The representative characteristics of the early stage of reprogramming are rapid proliferation6 and mesenchymal-to-epithelial transition (MET),7, 8 although the roles of these phenotypic signatures of reprogramming are controversial.9, 10 Gene expression profiles of the reprogramming process have identified several genes, such as Esrrb, Utf1 and EpCAM, that mark the initial phase.11, 12 However, expression of these genes is variable during the initial stage;11, 12 thus, it is unclear whether they can predict successful reprogramming.
The main obstacle to the study of the early reprogramming stage is its low efficiency,1, 2, 13, 14 which complicates elucidation of the mechanistic link between the reprogramming signal and final phenotype. In addition, the heterogeneity of somatic cells, such as progenitor cells versus terminally differentiated cells, may hinder elucidation of the genetic or epigenetic changes associated with this entire process as well as the molecular changes that occur during the initial stage.
We suggest that these obstacles can be overcome by using a defined reprogramming system with single cells that have an equal reprogramming potential. By generating this new system, we report that downregulation of extracellular signal-regulated kinase (ERK) and serum response factor (SRF) is a molecular signature of the early step of reprogramming.
Materials and methods
Cell culture
OG-MEFs and i4F-MEFs were isolated from B6;129S4-Pou5f1tm2Jae and R26rtTA;Col1al4F2A iPS mice (Jackson Laboratory, Sacramento, CA, USA) according to standard protocols and maintained in MEF medium (DMEM (#10-017-CVR); Corning) containing 10% fetal bovine serum (#16000-044; Thermo Fisher Scientific, Waltham, MA, USA) and 1% penicillin/streptomycin. 293FT and Plat-E cells were also cultured in MEF medium. J1 mESCs were purchased from ATCC (#SCRC-1010) and cultured on gelatin-coated dishes in standard mESC medium (DMEM supplemented with 15% fetal bovine serum (ESC grade, #S001-04; Welgene, Daegu, Korea), 1% penicillin/streptomycin, 2 mM glutamine, 0.1 mM nonessential amino acids, 1 mM β-mercaptoethanol and 103 units per ml LIF). Mouse iPSCs were cultured on mitomycin C-treated MEF feeder cells in mESC medium or KSR medium (mESC medium containing 15% KSR (Thermo Fisher Scientific) instead of fetal bovine serum).
Spontaneous immortalization of OG-MEFs
OG-MEFs were derived from OG2/ROSA26 heterozygous double-transgenic mice.15 These mice were generated by crossing the ROSA26 strain, which carries a neo/lacZ transgene, with the OG2 transgenic strain, which carries GFP under the control of the Oct4 promoter, over several generations. OG-MEFs at passage no. 5 were seeded at sub-confluency onto 60-mm dishes and kept in MEF medium for 3 weeks without medium replacement. Thereafter, the MEF medium was replaced every 7 days. After 2 months, cells were transferred to 100 mm dishes. After reaching confluency, cells were stored in nitrogen liquid at passage no. 7 or transferred to new dishes at a split ratio of 1:5. Cell culture was stopped at passage no. 40 without any observed changes in growth, survival or morphology.
Generation of reprogramming clones from siOG-MEFs
At passage no. 9, 2 × 104 siOG-MEFs were seeded into six-well plates. The following day, the lentiviral supernatant containing FUW-M2rtTA and Tet-O-FUW-OSKM was added to cells at a ratio of 1:10 (cell:virus) in the presence of polybrene (8 μg ml−1) for 12 h. The medium was replaced with fresh MEF medium, and the cells were incubated for 12 h. Thereafter, the cells were infected with a second aliquot of lentiviral supernatant at the same ratio for 12 h and maintained in fresh MEF medium for 3 days. After trypsinization, the cells were counted using a hemocytometer, seeded onto 96-well plates at a density of one cell per well and maintained for 3–4 weeks without medium replacement. Wells containing single colonies were observed using a light microscope and marked. Single colonies from marked wells were trypsinized and re-plated into two wells of a 24-well plate at a split ratio of 1:3 in the presence of 4 μg ml−1 Dox (25% of cells) or without Dox (75% of cells). After incubation for 3 days, untreated cells from the clones showing Dox-induced morphological changes (for example, MET and cell death) were collected and re-plated onto 60 mm dishes. After reaching confluency, the cells were transferred to 100-mm dishes. The cells were frozen at passage no. 1 or transferred to new dishes at a split ratio of 1:4 for subculture and reprogramming studies. These clones were named ‘JC’ because the vectors used were obtained from the laboratory of Dr Jaenisch (MIT). To generate other Dox-inducible stable reprogramming clones, a lentivirus encoding EF1α-TetR (#LVP-459-puro) and a Dox-inducible polycistronic cassette harboring OKSM (#LVP-359) were purchased from GeneTarget, Inc. (San Diego, CA, USA). LVP-359 does not contain any selection markers, such as antibiotic resistance or fluorescent proteins. Although this product is not commercially available, a similar product (#LVP381), which was not used in this study, harbors the same genes (in the order Myc, Klf4, Oct4 and Sox2) as well as the dual selection markers blasticidin resistance and red fluorescent protein. siOG-MEFs at passage no. 9 were infected with a lentivirus harboring EF1α-TetR at a ratio of 1:10 (cell:virus) and selected by treatment with puromycin (1 μg ml−1). siOG-MEFs stably expressing TetR were infected a second time with a lentivirus harboring OKSM at a ratio of 1:10 (cell:virus), and Dox-inducible stable reprogramming clones were generated according to the method described above. These clones were named ‘GC’ after the company.
Reprogramming efficiency and iPSC generation
To estimate the reprogramming efficiency using flow cytometry, cells were seeded into 100-mm dishes in the presence of Dox (4 μg ml−1). After incubation for 3 days, the cells were trypsinized and reseeded into 24-well plates at a density of 1 × 104 cells per well in the presence of Dox (Option I). The following day, MEF medium was washed away with PBS and replaced with E6 medium (#A15165-01; Thermo Fisher Scientific) containing LIF, Dox and chemical inhibitors. Alternatively, the cells were seeded into 12-well plates at a density of 2 × 104 cells per well. The following day, the cells were treated with Dox and incubated for an additional 3 days in MEF medium, which was then replaced with E6/LIF/Dox (Option II), or the cells were washed with PBS and directly placed in E6/LIF/Dox (Option III). The medium was replaced with fresh medium every 2 days. After incubation for 8 days, the cells were trypsinized, and GFP-positive cells were counted by flow cytometry. For the colony-formation assays, 400 cells were seeded onto MEF feeder cells in six-well plates. The following day, the cells were treated with Dox and maintained in MEF medium for 3 days. MEF medium was subsequently replaced with mESC/LIF/Dox, KSR/LIF/Dox or E6/LIF/Dox, and the cells were further incubated for 11 days. AP-positive colonies were detected using an AP detection kit (ScienCell Research Laboratories, San Diego, CA, USA), and GFP-positive colonies were manually counted by fluorescence microscopy. For iPSC generation, GFP-positive colonies (GC5, GC8, JC5, JC8 and JC17) grown in six-well plates were manually picked and reseeded onto MEF feeder cells. Among them, GC5-derived iPSCs were cultured in KSR medium containing LIF and CPT for chimera generation.
Phospho-kinase antibody array
GC5 cells were incubated in MEF medium containing Dox (4 μg ml−1) for 3 days and lysed with kinase lysis buffer (#ARY003B; R&D Systems, Minneapolis, MN, USA). Lysates (200 μg) were analyzed using a human phospho-kinase antibody array according to the manufacturer’s protocol (#ARY003B; R&D Systems). Membranes were analyzed using ImageJ software, National Institutes of Health, Bethesda, MD, USA (NIH).
Cell cycle analysis, immunocytochemistry and western blotting
Cell cycle analysis and immunocytochemistry were performed according to previously described methods.14, 16 For western blotting, cells were incubated with extraction buffer (#FNN0011; Thermo Fisher Scientific) to obtain total cell lysates and centrifuged at 16000 × g for 20 min. Western blot analysis was performed as previously described.3
siRNA. SRF-targeting (#1: sense, 5′-CCUAAACCACCUGUAUCCA-3′ and antisense, 5′-UGGAUACAGGUGGUUUAGG-3′ no. 2: sense, 5′-GGACGGCACCACUUAUUUA-3′ and antisense, 5′-UAAAUAAGUGGUGCCGUCC-3′ and no. 3: sense, 5′- GGUAGGGAGAUACAGGAAU-3′ and antisense, 5′-AUUCCUGUAUCUCCCUACC-3′) or control (#CP-004500-01-05) siRNA oligonucleotides were purchased from Bioneer. GC5 cells were transfected using DharmaFECT reagent (GE Healthcare, Marlborough, MA, USA) according to the manufacturer’s protocols. Cells in 12-well plates were transfected with 100 nM siRNA. The following day, cells were washed with PBS and the reprogramming assay was performed according to Option III. Alternatively, cells in six-well plates were transfected with 100 nM siRNA. The following day, the transfection medium was replaced with MEF medium containing Dox (4 μg ml−1). After incubation for 3 days, the cells were lysed for western blotting.
Luciferase reporter assay
Cells were infected with lentiviruses encoding SRE-luciferase (#CLS-010 L; Qiagen, Hilden, Germany) and Renilla (#CLS-RCL; Qiagen). After the indicated amount of time, the cells were washed with PBS and lysed with 5 × lysis buffer (Promega, Madison, WI, USA), and luciferase activity was measured according to the instructions for the Dual-Luciferase Reporter Assay System (Promega).
Reverse transcription PCR
Total RNA was extracted with TRIzol reagent (Thermo Fisher Scientific). cDNAs were synthesized from total RNA (2 μg) using 2 × cDNA Synthesis Master Mix (LeGene, San Diego, CA, USA), and PCR was performed with 2 × Direct MasterMix (Koma Biotech, Seoul, Korea). To quantitatively estimate gene expression, quantitative PCR was performed with 2 × SYBRGreen Mix (Koma Biotech), and the results were analyzed using a real-time PCR system (Bioneer, Daejeon, Korea). The PCR primers are listed in Supplementary Table 1.
ChIP assay
GC5 cells were infected with a lentivirus encoding Dox-inducible SRF-VP64-HA for 24 h. The medium was replaced with fresh MEF medium, and the cells were incubated for 24 h. Thereafter, the cells were infected with pMXs-retroviruses encoding DsRed, SRF and SRF-DN for 24 h and maintained in fresh MEF medium for 2 days. The cells were trypsinized and reseeded into 100-mm dishes. The following day, the cells were treated with Dox (4 μg ml−1) for 24 h and collected for the ChIP assay. ChIP was performed with an EZ-Magna Chip A/G kit (#17-10086; Millipore, Bedford, MD, USA) according to the manufacturer’s instruction. DNA was released from HA antibody-bound chromatin by reverse cross-linking and purified using spin columns. The PCR primers are listed in Supplementary Table 1.
Teratoma formation
Mouse iPSCs (1 × 106) were subcutaneously injected into 6-week-old BalB/c mice (n=4). Nine weeks after injection, teratomas were collected at the injection site, dissected and fixed overnight in 4% formaldehyde. They were then embedded in paraffin, sectioned at a thickness of 6 μm and analyzed by H&E staining. The experiments were approved by the Institutional Animal Care and Use Committee of CHA University and Chonbuk National University Medical School.
Aggregation of iPSCs with zona-free embryos and X-gal staining
Blastocysts were injected with iPSCs and stained with X-gal according to a previously described method.15 The experiments were approved by the Institutional Animal Care and Use Committee of Konkuk University.
Generation of chimeric mice
GC5-derived iPSCs were injected into ICR blastocysts and implanted into pseudopregnant ICR female mice. Chimeric mice were identified according to coat color. The experiments were approved by the Institutional Animal Care and Use Committee of Chonbuk National University Medical School.
Statistical analysis
Data are presented as the means±s.d. Statistical analysis was performed using two-tailed (or one-tailed), unpaired Student’s t-test for comparison of two samples. A P-value <0.05 was considered statistically significant. The number of samples examined is indicated by n.
Results
Generation of reprogramming clones from MEFs
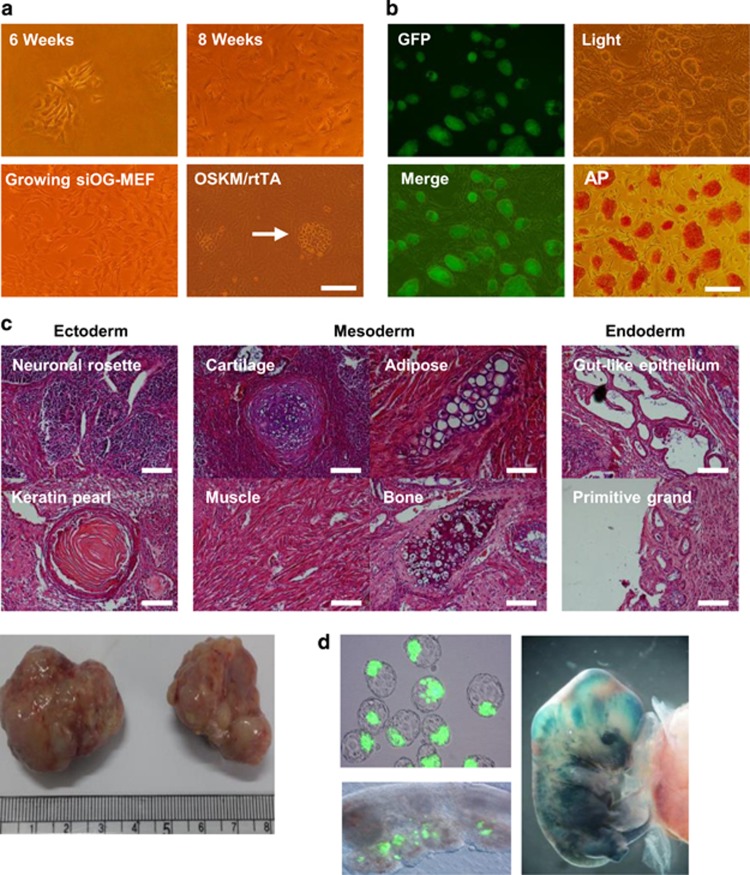
To generate reprogramming cell lines, we immortalized mouse embryonic fibroblasts (MEFs) with a reprogramming reporter system, namely, green fluorescent protein (GFP) expression driven by the ESC-specific promoter Oct4 (Oct4-GFP), which became growth-arrested within several passages in culture. When senescent Oct4-GFP-MEFs (OG-MEFs) were maintained in culture for approximately 4–6 weeks, some cells began to actively grow again (Figure 1a, upper panels) and continued to proliferate for over 40 passages without becoming senescent (Figure 1a, lower left panel). Spontaneously immortalized OG-MEFs (siOG-MEFs) exhibited MET (Figure 1a, lower right panel) upon enforced expression of reprogramming factors (Oct4, Sox2, Klf4 and Myc; OSKM) and then could be converted into induced pluripotent stem cells (iPSCs; Figure 1b). iPSCs formed teratomas in vivo (Figure 1c) and contributed to developing embryos in utero (Figure 1d). These data showed that siOG-MEFs had the potential to be fully reprogrammed by enforced expression of OSKM.
Figure 1.
siOG-MEFs reprogrammed by OSKM expression. (a) Morphology of siOG-MEFs. The arrow indicates Dox-induced MET of siOG-MEFs. Scale bar, 100 μm. (b) Generation of iPSCs from siOG-MEFs by Dox-inducible expression of OSKM. iPSCs were visualized as live cells (bright field and fluorescent images) or those stained for AP expression. Scale bar, 200 μm. (c) Teratoma formation by iPSCs. Hematoxylin and eosin (H&E) staining of teratoma sections showing the differentiation of mouse iPSCs into tissues of all three germ layers. Scale bars, 100 μm. (d) Embryo development and X-gal staining of iPSCs. iPSCs, induced pluripotent stem cells; MEFs, mouse embryonic fibroblasts; MET, mesenchymal-to-epithelial transition; siOG-MEFs, spontaneously immortalized OG-MEFs.
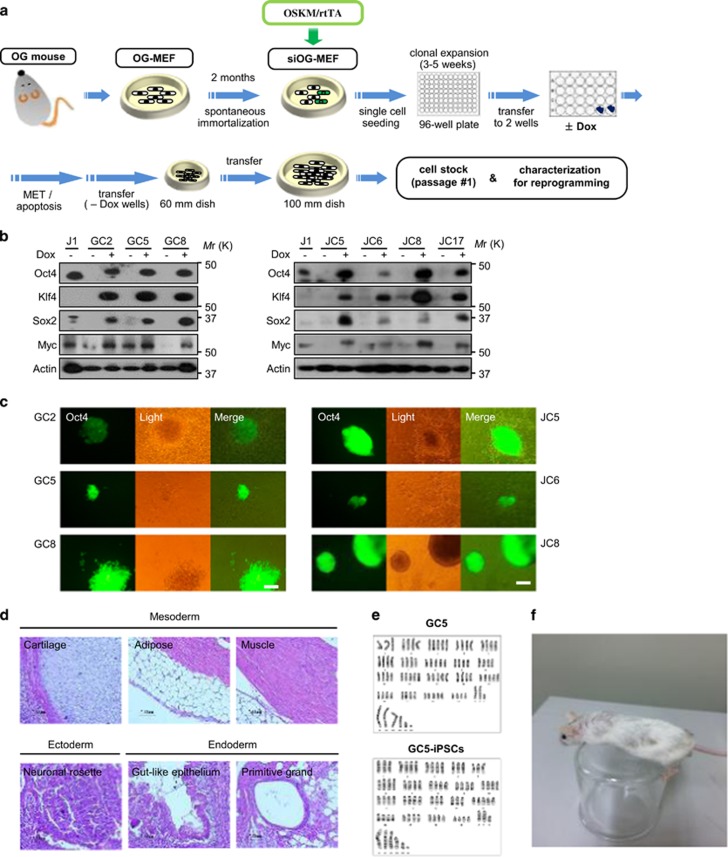
Next, siOG-MEFs were infected with a lentivirus encoding rtTA/OSKM or TetR/OKSM, which are Tet-On inducible vector systems for the expression of OSKM or OKSM from a single vector (Supplementary Figure la), and seeded into 96-well plates at a density of one cell per well (Figure 2a). Based on changes in cell morphology or survival induced by Doxycycline (Dox) treatment (4 μg ml−1), dozens of colonies were selected, and Dox-induced protein expression was examined by western blot analysis (Figure 2b and Supplementary Figure lb). Among them, GC5 and JC5 cells exhibited a MET phenotype within 3 days of culture in MEF medium containing Dox (Supplementary Figure 2a), developed an ESC-like morphology and expressed GFP when cultured in knockout serum replacement (KSR) medium containing leukemia inhibitory factor (LIF) and Dox (KSR/LIF/Dox; Figure 2c). These characteristics of both clones were conserved after conventional cell freezing and thawing.
Figure 2.
Dox-dependent reprogramming clones generated from siOG-MEFs. (a) Schematic for the generation of Dox-inducible mouse reprogramming clones. (b) Western blot analysis of the generated clones. Cells were treated with Dox (4 μg ml−1) for 3 days and collected for Western blot analysis. (c) Colony formation assay. Four hundred cells were seeded onto MEF feeder cells in six-well plates. The following day, cells were treated with Dox and maintained in MEF medium for 3 days. Thereafter, the MEF medium was replaced with KSR/LIF/Dox medium, and cells were further incubated for 11 days. Scale bars, 200 μm. (d) Teratoma formation by GC5-derived iPSCs. H&E staining of teratoma sections showing the differentiation of mouse iPSCs into tissues of all three germ layers. (e) Karyotype analyses showing 4 N abnormal chromosomes in GC5 cells and GC5-iPSCs. (f) Chimeric mouse generated from GC5-iPSCs. iPSCs, induced pluripotent stem cells; MEFs, mouse embryonic fibroblasts; MET, mesenchymal-to-epithelial transition; siOG-MEFs, spontaneously immortalized OG-MEFs.
Reprogramming cells are homogeneous and retain the ability to generate chimeras
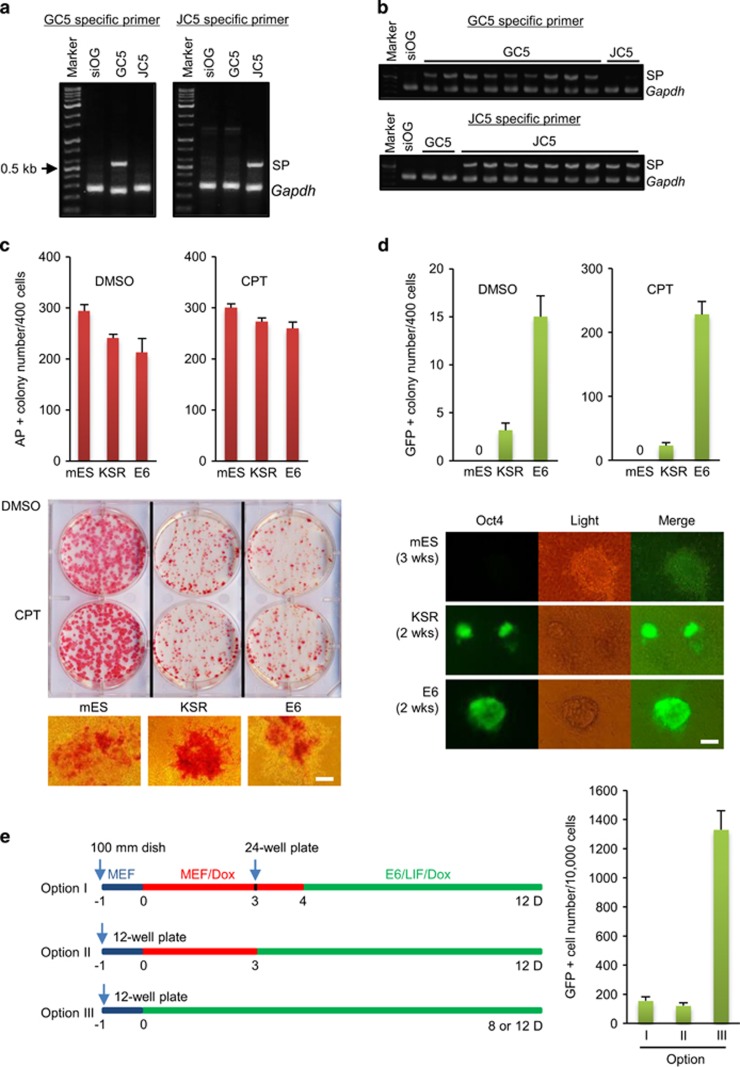
We examined whether the reprogramming clones were derived from single cells using GC5 and JC5 as representative clones because they showed a clear MET phenotype via E-cadherin upregulation (Figure 4d, E-cadherin panel, and Supplementary Figure 2c). Using chromosome walking analysis, we determined the insertion sites of the foreign genes in the GC5 and JC5 genomes and synthesized specific primers that were designed to amplify the foreign genes and host genome flanking sequences (Supplementary Figure 3a). PCR with genomic DNA revealed that the primer pairs were clone-specific (Figure 3a). The predicted PCR product was produced from nine primer-matched single cells, but not from single cells of parental siOG-MEFs or primer-mismatched clones (Figure 3b). Insertion of foreign DNA is stochastic; therefore, these data show that GC5 and JC5 cells originated from single cells. As expected, all cells of both clones expressed Oct4 upon Dox stimulation (Supplementary Figure 2b).
Figure 3.
Homogeneity and reprogramming efficiency of clones. (a) PCR analysis with genomic DNA. SP, specific product. (b) Single-cell PCR analysis. A single cell was isolated and directly subjected to PCR without DNA purification. (c, d) iPSC formation of GC5 cells. Cells (4 × 102) were seeded onto MEF feeder cells and incubated in the indicated medium containing CPT and Dox (4 μg ml−1). After 2 weeks, AP or GFP-expressing colonies were manually counted. Scale bars, 200 μm. (e) Time schedule for the reprogramming assay. The arrow indicates the time point of cell splitting and seeding. D, days; MEF, MEF medium containing 10% fetal bovine serum. The results are the means±s.d. of three independent experiments (two-tailed, unpaired t-test; n=2 (a, d) or 3 (e)). iPSCs, induced pluripotent stem cells; GFP, green fluorescent protein; MEFs, mouse embryonic fibroblasts; MET, mesenchymal-to-epithelial transition; OG-MEFs, Oct4-GFP-MEFs; siOG-MEFs, spontaneously immortalized OG-MEFs.
iPSCs generated from GC5 cells had pluripotency characteristics (Supplementary Figure 4a–c) and retained the ability to differentiate into all three germ layers in vitro (Supplementary Figure 4d) and in vivo (Figure 2d). However, GC5-derived iPSCs had an abnormal chromosome number of 4 N (Figure 2e and Supplementary Figure 4e) and failed to transmit through the germline during 6 months of observation (data not shown), although they formed chimeras at a high frequency (Figure 2f). Because GC5 cells can be reprogrammed for chimera generation, we further characterized this clone for use as a model system of induced pluripotency.
Effects of culture conditions and chemical compounds on GC5 cell reprogramming
When GC5 cells were cultured in mouse ESC (mESC) medium containing serum with LIF and Dox, almost all cells formed ESC-like colonies and expressed the ESC-specific marker alkaline phosphatase (AP; Figure 3c). However, none of the colonies expressed GFP (Figure 3d), implying that these colonies were not fully reprogrammed and that they resembled previously described pre-iPSCs.17 By contrast, culture in serum-free KSR medium or serum-free Essential 6 (E6) medium18 induced GFP expression (Figure 3d). This observation is consistent with previous reports that serum has an inhibitory effect on somatic cell reprogramming19, 20 and the final reprogramming of pre-iPSCs.17 The reprogramming efficiency under both culture conditions, as estimated by the formation of GFP-positive colonies, was as low as 1–3% of the seeded cells. These results suggest that the MET phenotype, as an early reprogramming signature, is necessary, but not sufficient, to induce reprogramming of somatic cells because almost all GC5 cells exhibited the MET phenotype (Supplementary Figure 2a) and growth stimulation (Supplementary Figure 2d) within 3 days of culture. During the development of an assay to reprogram the cloned cells, we observed that a temporal exposure (Options I and II) to serum upon initiation of reprogramming significantly decreased GFP-positive reprogramming in comparison with continuous culture under serum-free conditions (Option III; Figure 3e).
Some chemical inhibitors related to epigenetic and cellular signaling pathways can increase the reprogramming efficiency.21 Using GC5 cells, we observed that the MEK inhibitor PD0325901 (PD03), glycogen synthase kinase (GSK)-3β inhibitor CHIR99021 (CHIR) and histone deacetylase inhibitor trichostatin A (TSA) significantly increased the reprogramming efficiency (Supplementary Figure 3b). These results are consistent with previous reports using the retroviral system originally developed in Yamanaka’s laboratory.22, 23, 24 Notably, the reprogramming efficiency of cells exposed to all three compounds (1 μM CHIR, 0.1 μM PD03 and 0.01 μM TSA (CPT)) was ~80%, as estimated by flow cytometric analysis of GFP expression (Supplementary Figure 3c). In the colony formation assay, CPT treatment did not significantly affect colony formation or AP expression (Figure 3c), but induced GFP expression in all colonies grown under serum-free conditions (E6/LIF/Dox) (Figure 3d and Supplementary Movie 1). By contrast, almost all colonies expressed AP irrespective of the culture conditions and chemical treatment (Figure 3c), implying that the pre-iPSC stage in GC5 cells is a default process upon OSKM expression. Notably, AP (+)/GFP (−) colonies grown in serum-containing medium expressed GFP within 5 days of switching to serum-free medium (E6/LIF/Dox) containing CPT (data not shown). Collectively, these results suggest that synchronized metabolic (serum-free conditions plus CHIR plus PD03) and epigenetic (TSA) remodeling after the appearance of the MET phenotype is necessary for full reprogramming.
Signal transduction at an early stage of cellular reprogramming
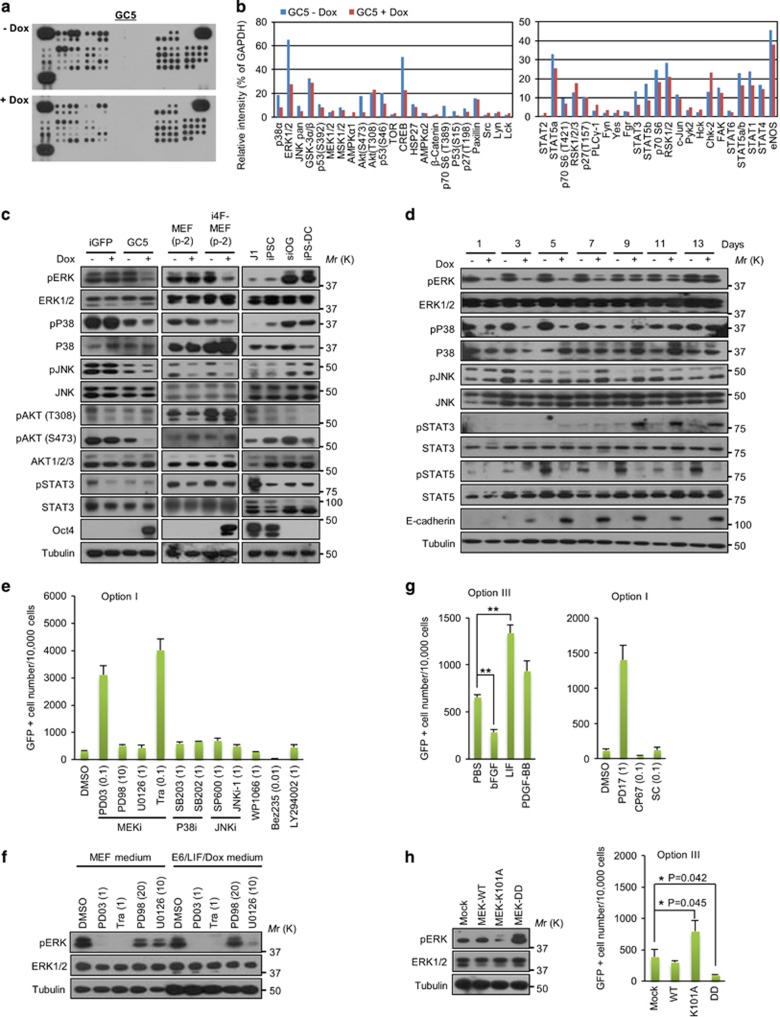
To explore the signal transduction pathways that act early in cellular reprogramming, Dox-treated or untreated GC5 cells were analyzed using a phospho-specific antibody array (Supplementary Figure 5). This analysis showed that phosphorylation of ERK, P38 and c-Jun N-terminal kinase (JNK) was significantly lower in Dox-treated cells than in untreated cells (Figures 4a and b). These results were confirmed by Western blot analysis of GC5 cells (Figure 4c, left) and Dox-inducible normal i4F-MEFs (Figure 4c, middle) isolated from iPS mice.25 When mitogen-activated protein (MAP) kinase signaling was examined in Dox-treated or untreated GC5 cells for 13 days, ERK and p38 signaling were downregulated during the early/intermediate stages of reprogramming and recovered in the late stage (Figure 4d). ERK signaling was lower in mouse iPSCs than in their parental somatic cells, siOG-MEFs and cells differentiated from iPSCs (Figure 4c, right). These data indicate that suppression of ERK signaling is an early event in reprogramming signal transduction and that this pathway is downregulated for iPSC formation.
Figure 4.
ERK suppression is an early signal of cellular reprogramming and increases its efficiency. (a) Phospho-kinase antibody array with GC5 cells incubated in MEF medium containing Dox (4 μg ml−1) for 3 days. (b) Relative intensities of the phospho-kinase spots. Membranes were analyzed using ImageJ software (NIH). (c) Western blot analysis. Cells were incubated in MEF medium containing Dox (4 μg ml−1) for 3 days. Alternatively, cells were cultured in fresh mESC medium containing LIF (J1 mESCs and iPSCs) or in fresh MEF medium (siOG-MEFs and cells differentiated from iPSCs). i4F-MEF, Dox-inducible OSKM-expressing normal MEFs; P-2, passage no. 2; iPS-DC, cells differentiated from iPSCs. (d) Western blot analysis with GC5 cells incubated in MEF medium containing Dox (4 μg ml−1) for the indicated number of days. (e) Reprogramming assay with chemical inhibitors against MAP kinases and some signaling molecules. Sub-lethal concentrations of the chemicals were determined by testing various concentrations (0.01, 0.1, 1 and 10 μM) using Option I (μM). MEK inhibitors: PD03, PD98 (PD98059), U0126 and Tra (trametinib). P38 inhibitors: SB203 (SB203580) and SB202 (SB202190). JNK inhibitors: SP600 (SP600125) and JNKi-1 (JNK inhibitor I). STAT3 inhibitor: WP1066. mTOR inhibitor: Benz235. PI3K inhibitor: LY294002. (f) Western blot analysis of pERK expression in GC5 cells incubated with the indicated chemicals and medium for 24 h. (g) Reprogramming assay of GC5 cells with cytokines (Option III; concentration, 10 ng ml−1) or chemicals (Option I; μM). PD17 (PD173074, FGFR inhibitor), CP67 (CP673451, STAT3 inhibitor) and SC (SC144, PDGFR inhibitor). (h) Western blot analysis of pERK expression and reprogramming assay in GC5 cells infected with a retrovirus encoding MEK-WT, MEK-K101A or MEK-DD. After 3 days of infection, cells were subjected to Western blot analysis or were detached and seeded onto 12-well plates for the Option III assay. The results are the means±s.d. of three independent experiments (two-tailed, unpaired t-test; n=2 (e) or n=3 (g,h)). *P<0.05; **P<0.01. ERK, extracellular signal-regulated kinase.
ERK suppression is a driving force for cellular reprogramming
To examine whether inhibition of MAP kinases is functionally linked to cellular reprogramming, GC5 cells were treated with various chemical compounds that inhibit MAP kinases and other signaling kinases. Using MEK inhibitors with different IC50 values, two compounds with low IC50 values (PD03: IC50=0.33 nM and trametinib: IC50=0.92 nM) dramatically increased the reprogramming efficiency at a sub-lethal concentration (0.1 μM) under serum-free reprogramming conditions (Figure 4e) and potently inhibited phosphorylation of ERK (Figure 4f). However, PD98059, which has a high IC50 value (2 μM), had little effect on the reprogramming efficiency and ERK phosphorylation (Figures 4e and f). U0126, which has a moderate IC50 value (70 nM), significantly inhibited ERK phosphorylation at a high concentration (10 μM), but the reprogramming efficiency was low at a sub-lethal concentration (1 μM; Figures 4e and f). P38 and JNK inhibitors increased the reprogramming efficiency (Figure 4e), but these effects were small in comparison with those of MEK inhibitors with low IC50 values.
Basic fibroblast growth factor (bFGF) strongly activates ERK through its receptor fibroblast growth factor receptor (FGFR).26 As expected, bFGF reduced the reprogramming efficiency, while a chemical inhibitor of FGFR strongly increased it (Figure 4g). Because mESCs lose pluripotency when ERK is activated,27, 28 these data indicate that downregulation of ERK signaling drives cellular reprogramming. To further examine this possibility, GC5 cells were infected with a retrovirus encoding a dominant-negative MEK (MEK/K101A) to decrease ERK phosphorylation or a constitutively active MEK (MEK/DD) to increase ERK phosphorylation. In comparison to control cells, MEK/K101A-expressing cells had an increased reprogramming efficiency, while MEK/DD-expressing cells had a decreased reprogramming efficiency (Figure 4h). These data demonstrate that downregulation of ERK signaling is a driving force for cellular reprogramming.
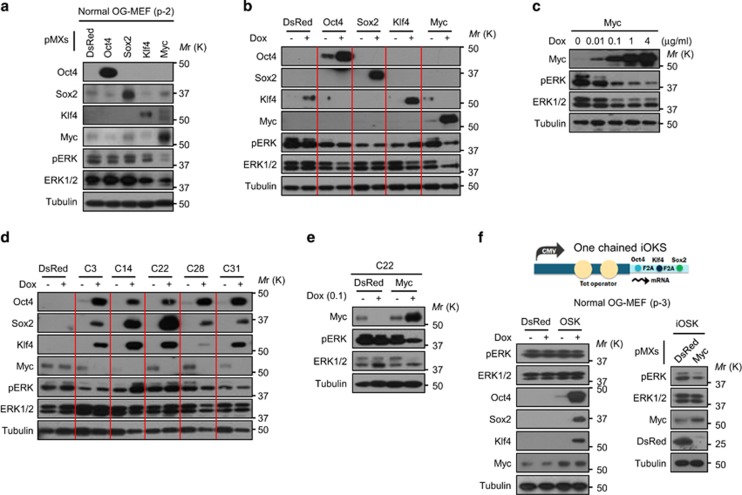
Myc inhibits ERK signaling in immortalized and normal MEFs
To identify the reprogramming factors responsible for ERK inhibition, normal OG-MEFs were infected with retroviruses encoding these factors. ERK phosphorylation was greatly reduced in Myc-expressing cells (Figure 5a). This phenomenon was also observed in Myc-expressing siOG-MEFs (Figure 5b). Indeed, Myc suppressed ERK phosphorylation in a dose-dependent manner (Figure 5c). We examined whether the combined reprogramming factors, excluding Myc, could inhibit ERK phosphorylation as a cooperative effect. To achieve this goal, clones simultaneously expressing OSK upon Dox treatment were isolated from siOG-MEFs. The selected clones did not show a reduction of ERK phosphorylation upon Dox treatment (Figure 5d), but showed reduced ERK phosphorylation in the presence of Myc (Figure 5e). We obtained a similar result in normal OG-MEFs expressing OSK at an equal molar ratio (Figure 5f). Myc greatly boosts the efficiency of cellular reprogramming, although it is not absolutely necessary for reprogramming.29, 30 Therefore, our results suggest that Myc can facilitate cellular reprogramming through, at least in part, suppression of ERK signaling.
Figure 5.
Myc inhibits ERK signaling in MEFs. (a) Western blot analysis of pERK and reprogramming factor expression in normal OG-MEFs (passage no. 2) separately infected with retroviruses encoding reprogramming factors or DsRed for 4 days. pMXs, pMXs-retrovirus. (b) Western blot analysis of pERK and reprogramming factor expression in siOG-MEFs separately infected with lentiviruses encoding Dox-inducible reprogramming factors or DsRed after 3 days of Dox treatment. (c) Western blot analysis of pERK and Myc expression in the Dox-inducible Myc expressing siOG-MEFs treated with various concentrations of Dox for 3 days. (d) Western blot analysis of pERK and reprogramming factor expression in the iOSK-expressing siOG-MEF clones separately infected with lentiviruses encoding Dox-inducible Oct4, Sox2 and Klf4. Cells were treated with Dox (4 μg ml−1) for 3 days. (e) Western blot analysis of pERK and Myc expression in the iOSK-expressing clone C-22 infected with a lentivirus encoding Dox-inducible DsRed or Myc for 24 h. Cells were treated with Dox (0.1 μg ml−1) for 3 days. (f) Western blot analysis of pERK and reprogramming factor expression in normal OG-MEFs (passage no. 3) infected with a lentivirus encoding Dox-inducible OKS as a single polycistronic unit. Cells were treated with Dox (4 μg ml−1) for 3 days. Alternatively, after co-infection with a retrovirus encoding DsRed or Myc, cells were treated with Dox (4 μg ml−1) for 5 days. ERK, extracellular signal-regulated kinase; OG-MEFs, Oct4-GFP-MEFs.
SRF suppression enhances iPSC formation in GC5 cells
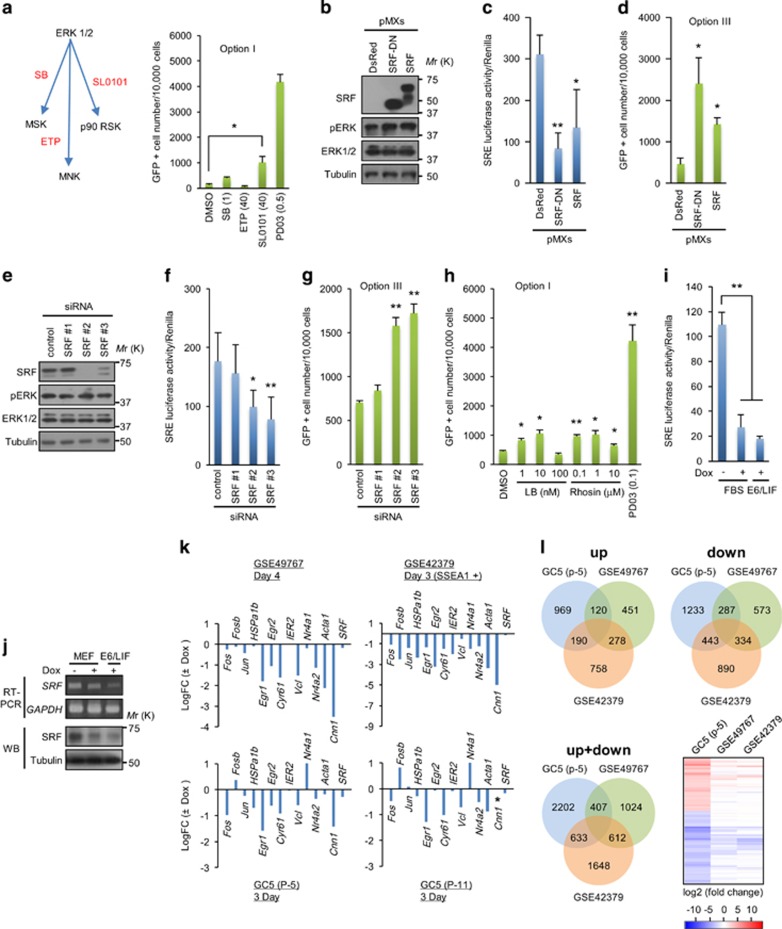
To elucidate how ERK signaling is able to modulate cellular reprogramming, we explored target molecules downstream of ERK using GC5 cells. We focused on ERK downstream kinases, such as MAP kinase-interacting kinases, mitogen- and stress-activated kinase and 90 kDa ribosomal S6 kinases (RSKs), because they are essential to transduce ERK signaling to their multiple target molecules31 and chemical inhibitors of these kinases are available (Figure 6a, left). The RSK inhibitor significantly increased the efficiency of reprogramming in GC5 cells (Figure 6a, right), although its effect was much less than that of the MEK/ERK inhibitor.
Figure 6.
SRF suppression increases the efficiency of reprogramming and is a molecular sign of an early reprogramming event. (a) Kinases downstream of ERK and their chemical inhibitors (left panel). SB, SB747651A; ETP, ETP45835. The assay was performed according to Option I (right panel). (b) Western blot analysis of SRF and pERK expression in GC5 cells infected with retroviruses encoding DsRed, SRF-DN and SRF. (c) Luciferase assay of GC5 cells infected with lentiviruses encoding SRE-luciferase and Renilla and then with retroviruses encoding DsRed, SRF-DN and SRF for 3 days. Luciferase activity was measured in cell lysates and normalized to Renilla activity in the same sample. (d) Reprogramming assay of GC5 cells expressing DsRed, SRF-DN and SRF. (e) Western blot analysis of SRF and pERK expression in GC5 cells transfected with control or SRF-targeting siRNA oligonucleotides. (f) Luciferase assay of GC5 cells infected with lentiviruses encoding SRE-luciferase and Renilla. The luciferase assay was performed 3 days after transfection for siRNA. (g) Reprogramming assay of GC5 cells transfected with control or SRF-targeting siRNA oligonucleotides. (h) Reprogramming assay of GC5 cells treated with chemicals. (i) Luciferase assay of the SRF-reporter expressing GC5 cells after Dox treatment for 3 days. (j) Western blot analysis and reverse transcription PCR (RT-PCR) of SRF expression in GC5 cells treated with Dox (4 μg ml−1) for 3 days. (k) RNA-seq analysis of GC5 cells (passage no. 5 and no. 11) treated with Dox (4 μg ml−1) for 3 days. Fold changes in expression of the selected IEGs are presented as the log2FC (fold change). Log2FC of IEGs in GC5 cells was compared with those of IEGs from two published datasets (GSE49767 and GSE42379). (l) The similarity of the transcriptional profiles of GC5 cells (passage no. 5) and the GSE49767 and GSE42379 datasets was determined according to the number of genes that were commonly upregulated or downregulated by Dox (total genes=9404). The heat map presents the normalized expression levels (log2FC) with red and blue representing up- and downregulated genes, respectively. GC5, RNA-seq data from GC5 cells. The results are the means±s.d. of three independent experiments (two-tailed, unpaired t-test; n=2 (h) or n=3 (a, c, f, i)). *P<0.05; **P<0.01. ERK, extracellular signal-regulated kinase; IEGs, immediate-early genes; SRF, serum response factor.
RSKs regulate biological processes, such as cell proliferation and survival, through phosphorylation of their target substrates.32 Among them, the transcription factor SRF regulates many genes through the CArG box, a core component of the serum response element (SRE),33 and is essential for mesoderm development.34 This protein can also be activated by ERK-induced phosphorylation of members of the ternary complex factor (TCF) subfamily (Elk-1, Elk-3 and Elk-4), the binding site of which is adjacent to the CArG box within the SRE.31, 33 To simply test whether SRF is involved in reprogramming modulation, we used a chemical inhibitor of this molecule (CCG1423). CCG1423 treatment not only suppressed SRF transcriptional activity in GC5 cells (Supplementary Figure 6a), but also significantly increased their reprogramming (Supplementary Figure 6b). However, CCG1423 potently inhibited ERK phosphorylation in GC5 cells under serum-free conditions (Supplementary Figure 6a), indicating that this compound was not highly target-specific.
To suppress SRF activity with an improved specificity, we generated a mutant SRF with a truncated activation domain. This SRF mutant can block SRF function in a dominant-negative manner.35 Dominant-negative SRF (SRF-DN) reduced SRF activity without affecting ERK phosphorylation (Figures 6b and c) and enhanced the efficiency of reprogramming (Figure 6d). Next, we examined the effect of loss-of-function of SRF on cellular reprogramming using three different SRF-targeting small interfering RNAs (siRNAs). Two siRNAs (no. 2 and no. 3) significantly downregulated SRF expression in GC5 cells without any effect on ERK phosphorylation, and they also decreased SRF activity in these cells (Figures 6e and f). Both SRF-targeting siRNAs increased the efficiency of reprogramming (Figure 6g). SRF can be activated by ERK-mediated TCF phosphorylation or RhoA/actin-mediated MKL1 activation.31 The MKL1 inhibitors latrunculin B and rhosin increased the reprogramming efficiency, but these effects were much smaller than that of the MEK inhibitor (Figure 6h). These data indicate that suppression of ERK-SRF signaling is important for increasing reprogramming.
SRF is suppressed at an initial phase of cellular reprogramming
Because SRF is downstream of ERK in MAP kinase signaling cascades, this protein may be concomitantly suppressed by ERK during the reprogramming process. To test this possibility, we induced reprogramming for 3 days in GC5 cells expressing a SRF reporter gene. Measurement of luciferase activity demonstrated that SRF transcriptional activity was downregulated (Figure 6i; note that GC5 cells could not survive in the absence of Dox under serum-free conditions). SRF strongly induces immediate-early genes (IEGs) and belongs to the IEG family.34, 35 Therefore, expression of SRF was also downregulated at an early stage of GC5 cell reprogramming (Figure 6j). RNA-seq analysis revealed that expression of other IEGs beside SRF was decreased at this stage of reprogramming (Figure 6k). We obtained a similar result by analyzing cDNA array data, which represent the gene expression profiles of normal i4F-MEFs at the onset of reprogramming (day 3 or 4), as published by two other groups12, 36 (Figure 6k and Supplementary Table 2). Interestingly, the gene expression profiles of reprogramming GC5 cells correlated with those of normal i4F-MEFs (Figure 6l and Supplementary Table 3). These data show that SRF is downregulated at an early step of cellular reprogramming.
IEGs other than Myc do not promote reprogramming
The reprogramming factor Myc can be upregulated by the ERK pathway37 and belongs to the IEG family. To test whether other IEGs can regulate cellular reprogramming similar to Myc, we infected normal OG-MEFs with retroviruses encoding several IEGs, including c-Jun, and examined their effects on ERK phosphorylation. Myc inhibited ERK phosphorylation, whereas c-Fos and c-Jun increased ERK phosphorylation (Supplementary Figure 7a). In the presence of OSK at an equal molar ratio, retrovirally expressed Myc significantly increased the reprogramming of normal OG-MEFs, whereas other IEGs (SRF, c-Fos, c-Jun and EGR1) had little effect (Supplementary Figure 7b and c). Thus, the IEGs examined could not replace the role of Myc in cellular reprogramming.
SRF is essential for modulation of somatic cell reprogramming by ERK
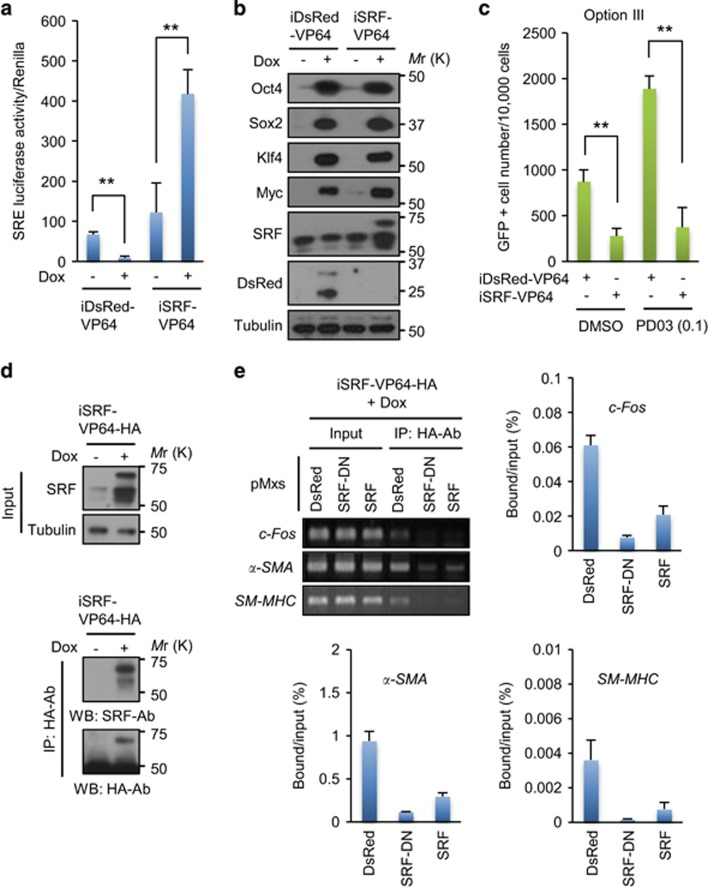
To determine whether ERK is coupled to SRF at the molecular level for reprogramming modulation, we examined the effect of SRF activation on reprogramming induced by ERK inhibition. If ERK inhibition increased reprogramming via SRF suppression, SRF activation would suppress it. For this experiment, we did not use the SRF overexpression system because it unexpectedly decreased SRF transcriptional activity (Figure 6c). Similarly, it has been reported that overexpression of SRF does not induce expression of its target genes.38 SRF transcriptional activity can be finely regulated by the presence of multiple cofactors, including members of the TCF subfamily and myocardin-related factors (i.e., MKL1).33, 39 These cofactors can also be activated by external stimuli such as serum and growth factors.33 Therefore, it is possible that SRF overexpression is insufficient to induce its activity without external stimulation.
To overcome this problem, we generated a mutant form of SRF (SRF-VP64) that has a VP-64 domain within its C-terminal region. The transactivation domain VP-64 can activate transcription factors by recruiting their co-activators without external stimuli.40 As expected, when Dox-inducible SRF-VP64 (iSRF-VP64) or DsRed-VP64 (iDsRed-VP64) was expressed in GC5 cells, SRF-VP64 protein blocked reprogramming-induced SRF suppression and greatly boosted SRF activity, while DsRed-VP64 had no effect (Figure 7a). SRF-VP64 did not alter OSKM expression in these cells (Figure 7b). Importantly, SRF-VP64 not only decreased GC5 cell reprogramming but also suppressed the reprogramming induced by ERK inhibition (Figure 7c). To determine the target specificity of SRF-VP64, we performed chromatin immunoprecipitation (ChIP) using the SRE region within the promoters of c-Fos, α-smooth muscle actin (α-SMA) and smooth muscle myosin heavy chain (SM-MHC), which are target genes of SRF.41 To achieve this goal, we generated the HA-tagged iSRF-VP64 vector and confirmed that an anti-HA antibody successfully precipitated the SRF-VP64-HA protein from GC5 cells (Figure 7d). ChIP experiments revealed that the binding of the SRF-VP64-HA protein to these SRE regions was blocked in the presence of SRF or SRF-DN (Figure 7e), indicating that SRF-VP64 was activated through an interaction with SRE regions. Taken together, these data show that SRF is a target molecule of ERK for reprogramming modulation.
Figure 7.
SRF activation suppresses reprogramming induced by ERK inhibition. (a) Luciferase assay of the SRF-reporter expressing GC5 cells infected with a lentivirus encoding iDsRed-VP64 or iSRF-VP64 after Dox treatment for 3 days. (b) Western blot analysis of SRF and reprogramming factor expression in a. (c) Reprogramming assay of GC5 cells in (A). After 24 h of infection with a lentivirus encoding iDsRed-VP64 or iSRF-VP64, MEF medium was changed to E6/LIF/Dox medium with or without PD03 (0.1 μM). (d) Western blot analysis of SRF expression in GC5 cells infected with a lentivirus encoding iSRF-VP64-HA. After infection, cells were treated with or without Dox (4 μg ml−1). After incubation for 3 days, cells were lysed with kinase lysis buffer (R&D Systems). (e) ChIP assay for IEG promotor binding of SRF-VP64. GC5 cells were infected with a lentivirus encoding iSRF-VP64-HA for 24 h, replenished with fresh MEF medium and separately infected with retroviruses encoding DsRed, SRF-DN and SRF. Data are representative of two independent experiments and the mean of triplicates (per biological sample). In a and c, the results are the means±s.d. of three independent experiments (two-tailed, unpaired t-test; n=3). **P<0.01. ERK, extracellular signal-regulated kinase; IEGs, immediate-early genes; IP, immunoprecipitation; SRF, serum response factor.
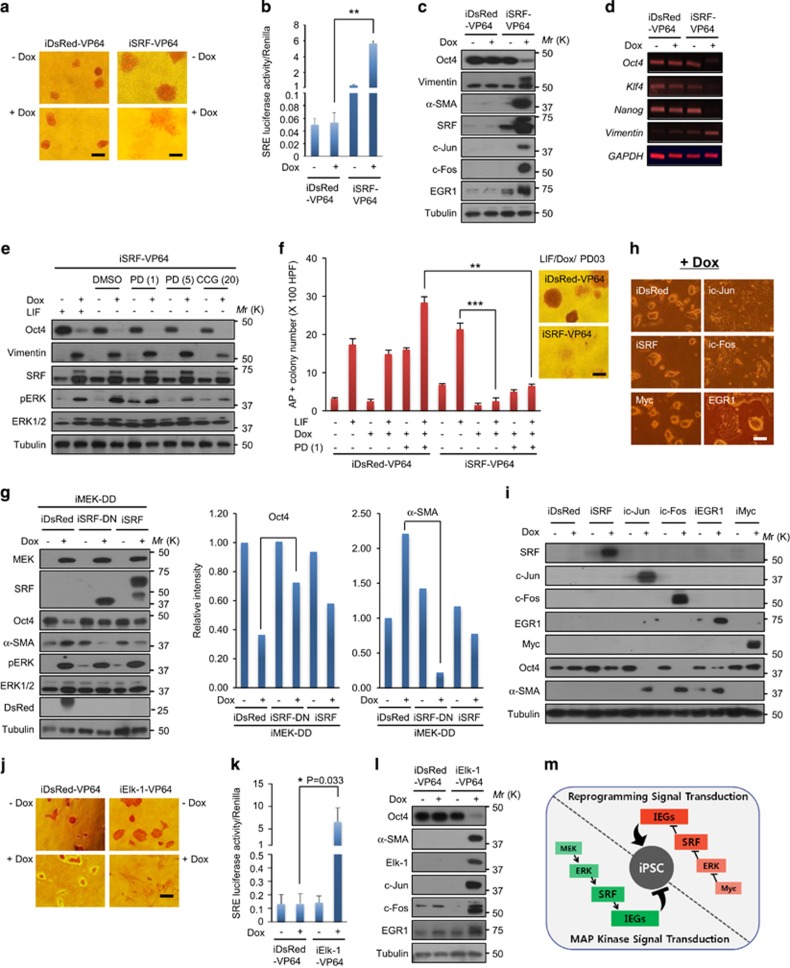
SRF activation negatively regulates embryonic pluripotency
To determine how SRF activation impedes somatic cell reprogramming, we examined its effect on embryonic pluripotency. We reasoned that SRF activation would promote stem cell differentiation. Indeed, when infected with a lentivirus encoding iSRF-VP64 and then treated with Dox, mESCs underwent marked morphological changes that were indicative of differentiation (Figure 8a) and demonstrated enhanced SRF activity (Figure 8b). This phenotypic change was not observed in mESCs infected with a lentivirus encoding iSRF (Supplementary Figure 8a). However, these cells showed significantly increased SRF activity in the presence of Dox, but their total SRF activity was approximately 80-fold lower than that in cells expressing SRF-VP64 (Supplementary Figure 8b). Because SRF expression was much lower in mESCs than in somatic cells (Supplementary Figure 8d), SRF activity increased by a lentivirus encoding iSRF as well as its basal SRF activity may be insufficient to drive the differentiation of mESCs.
Figure 8.
SRF activation negatively regulate pluripotency in mESCs through IEG induction. (a) AP expression in J1 mESCs infected with a lentivirus encoding iDsRed-VP64 or iSRF-VP64. After infection, cells were treated with or without Dox (4 μg ml−1) for 2 days. Scale bars, 100 μm. (b) Luciferase assay of the SRF reporter-containing J1 mESCs in a. (c) Western blot analysis of Oct4 and IEG expression in b. (d) reverse transcription PCR (RT-PCR) analysis of vimentin and stemness factor expression in a. (e) Western blot analysis. J1 mESCs were infected with a lentivirus encoding iSRF-VP64 for 24 h and incubated in fresh mESC medium containing LIF for 2 days. After replacement of the medium with mESC medium without LIF, cells were treated with PD03 or CCG1423 for 1 h before adding Dox. After incubation for 2 days with or without Dox (4 μg ml−1), cells were collected for western blotting. (f) AP expression of J1 mESCs in e. AP-positive colonies were counted in 100 high-power fields (HPFs) under a light microscope. Scale bar, 100 μm. (g) Western blot analysis. J1 mESCs were infected with a lentivirus encoding iMEK-DD for 24 h and then with lentiviruses encoding iDsRed, iSRF-DN and iSRF. After incubation for 2 days with or without Dox (4 μg ml−1), cells were collected for Western blotting. The intensities of the protein bands were analyzed using ImageJ software (NIH). (h) Morphological changes in J1 mESCs infected with a lentivirus encoding iDsRed or Dox-inducible IEGs such as c-Jun, c-Fos, EGR1, Myc and SRF. After infection, cells were treated with or without Dox (4 μg ml−1) for 2 days. Scale bar, 100 μm. (i) Western blot analysis of IEG and α-SMA expression in h. (j) AP expression in J1 mESCs infected with a lentivirus encoding iDsRed-VP64 or iElk-1-VP64. After infection, cells were treated with or without Dox (4 μg ml−1) for 2 days. Scale bars, 100 μm. (k) Luciferase assay of the SRF reporter-containing J1 mESCs in (j). (l) Western blot analysis of Oct4 and IEG expression in j. (m) Diagram showing the proposed reprogramming signal transduction pathway. The results are the means±s.d. of three independent experiments (one-tailed (k) or two-tailed (b, f), unpaired t-test; n=2 (f) or n=3 (b, k)). *P<0.05; **P<0.01; ***P<0.001. IEGs, immediate-early genes; mESCs, mouse embryonic stem cells; SRF, serum response factor.
By contrast, SRF-VP64 decreased Oct4 expression and increased expression of the mesoderm differentiation markers α-SMA and vimentin (Figures 8c, d and Supplementary Figure 8e). Again, SRF failed to induce Oct4 downregulation and α-SMA upregulation in mESCs (Supplementary Figure 8c). SRF-VP64 induced upregulation of the EMT genes Snail and Slug as well as downregulation of several pluripotency genes, including Nanog in mESCs (Supplementary Figure 8e). Because iPSC formation is facilitated by upregulation of pluripotency genes and suppression of EMT genes, these data indicate that SRF activation impedes somatic cell reprogramming. Interestingly, induction of mESC differentiation by SRF-VP64 significantly enhanced ERK phosphorylation (Figure 8e). However, suppression of ERK phosphorylation by the MEK inhibitor failed to block mESC differentiation induced by SRF-VP64 (Figure 8f), suggesting that SRF is downstream of ERK in pluripotency modulation. To validate this hypothesis, we induced ERK phosphorylation and differentiation in mESCs by using a lentivirus encoding a constitutively active MEK (iMEK/DD). We found that SRF-DN inhibited the MEK/DD-induced differentiation in mESCs without alternating ERK phosphorylation (Figure 8g), showing that ERK regulates pluripotency through SRF.
Finally, we examined the effect of IEGs on embryonic pluripotency. When mESCs were infected with lentiviruses encoding Dox-inducible IEGs, c-Jun and c-Fos strongly induced the differentiation of these cells, EGR1 moderately induced their differentiation, whereas SRF and Myc had no effect (Figures 8h and i). As expected, the constitutively active form of SRF (SRF-VP64) induced expression of IEGs, such as c-Jun, c-Fos and EGR1, in mESCs (Figure 8c). The effect of SRF-VP64 on mESC differentiation was reproduced by using the constitutively active form of Elk-1 (Elk-1-VP64), which directly received the signal for SRF activation from ERK (Figures 8j–l). These data indicate that SRF activation suppresses the pluripotency of mESCs through upregulation of IEGs.
Discussion
We explored the possibility of cell line-based studies for cellular reprogramming in stem cell biology. Using a defined system with an equal reprogramming potential, this study revealed several new concepts for somatic cell reprogramming.
The first finding is the existence of a reprogramming barrier at a late stage. Using a homogeneous single cell-derived population, the MET phenotype was induced in almost 100% of cells, showing that all cells entered the early stage of reprogramming. Almost all cells formed ESC-like colonies and expressed the ESC-specific marker AP irrespective of the culture conditions. However, none of these cells could be fully reprogrammed (Oct4-GFP-positive) when cultured in serum-containing medium, implying that they were partially reprogrammed. Thus, these cells had an equal reprogramming potential until the pre-iPSC stage. These data indicate that serum can hamper reprogramming in the final stage, but not in the pre-iPSC state.
The second finding is that metabolic and epigenetic remodeling is needed to overcome the reprogramming barrier. Under serum-free conditions, cells could be fully reprogrammed, but the reprogramming efficiency was still low in the absence of small-molecule inhibitors (CPT). With a combination of serum-free medium and CPT, almost all cells were fully reprogrammed. These two findings are similar to the results obtained for the conversion of pre-iPSCs into iPSCs.17 This previous study demonstrated that pre-iPSC colonies with extremely weak expression of Oct4-GFP are generated by retroviral transduction of OSKM in the presence of serum and LIF, and many of these colonies are converted into GFP-positive iPSCs when switched to serum-free medium containing 2i (PD03 and CHIR) and LIF. Together with this previous report, our findings may provide the optimal experimental conditions to achieve a high reprogramming efficiency.
The third finding is that ERK suppression is an early molecular event for reprogramming facilitation. We demonstrated that Myc was responsible for inhibition of ERK signaling during reprogramming. Myc can support the pluripotency of mESCs through ERK suppression.42 Thus, Myc-induced ERK suppression may be favorable to reprogram cells into the ESC state. Interestingly, ERK signaling was downregulated during the early/intermediate stages of reprogramming and recovered at the late stage. By contrast, STAT3, which is a key molecule mediating the self-renewal and pluripotency of mESCs,43, 44 was activated during the intermediate/late stages (Figure 4d). Because ERK signaling was remarkably lower in pluripotent cells than in somatic cells, the former finding suggests the existence of a molecular defense system against reprogramming in somatic cells. Consistent with this observation, Niwa et al.45 reported that inhibition of ERK signaling promotes the pluripotency of mESCs through upregulation of Nanog and Tbx5. This defense system may explain why some potent MEK/ERK inhibitors dramatically increase the reprogramming efficiency.
The fourth finding is that SRF is downstream of ERK for reprogramming modulation. ERK regulates several biological processes, such as proliferation, survival and differentiation.26, 46 To mediate these events, ERK can directly or indirectly activate many target molecules.31, 46 This variability in ERK signaling raises the question of whether ERK is a final executor of reprogramming modulation. To address this possibility, we explored the effect of target molecules downstream of ERK on induced pluripotency. Using our reprogramming clone, we observed that inhibition of SRF increased the reprogramming efficiency and that this molecule was suppressed at an early stage of reprogramming. Furthermore, reprogramming induced by ERK inhibition was impaired by SRF activation. SRF activation negatively regulated embryonic pluripotency. Subsequently, we showed that IEGs, such as c-Jun, c-Fos and EGR1, induced differentiation of mESCs. Because IEGs are downstream of SRF, these data indicate that SRF can modulate embryonic pluripotency through IEGs. c-Jun induces differentiation of mESCs through upregulation of differentiation-related genes (Slug and Snail) and downregulation of pluripotency genes (Oct4 and Sox2).47 Together with this previous report, the present study explains why SRF should be suppressed prior to completion of reprogramming. We hypothesize that suppression of the ERK-SRF-IEG axis, which is a MAP kinase signaling pathway, transduces a reprogramming signal for iPSC formation (Figure 8m). By contrast, it has been reported that ERK negatively regulates embryonic pluripotency by inducing destabilization of Klf2.48 Hence, it is possible that ERK modulates embryonic pluripotency through a number of molecules including Klf2 and SRF. However, it remains to be determined whether ERK modulates somatic cell reprogramming via Klf2.
Although we demonstrated the inhibitory effect of SRF on induced pluripotency for the first time, this finding may be intuitively inferred from its positive role in development and its hierarchical position within the ERK signaling cascade. Nevertheless, our finding that suppression of the ERK-SRF axis is an early molecular event in somatic cell reprogramming may be of particular interest in the stem cell and developmental biology fields. mESCs are derived from the inner cell mass49 and retain a full developmental potential, termed ‘naive’ pluripotency, when cultured in vitro.50 The key characteristic feature of murine ESCs is their propagation in the absence of ERK signaling.26, 50 The cell-autonomous capacity to thrive independently of ERK arises during blastocyst development and is rapidly lost after implantation.51 Together with these developmental studies, the present study suggests that the erasure of ERK signaling (or ERK-SRF signaling) is a prerequisite for somatic cells to return to the naïve state. Suppression of the ERK-SRF axis may be a useful surrogate marker for the development of new reprogramming technologies and for classical reprogramming methodologies such as gene transfer, nuclear transfer and cell fusion.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
This research was supported by the Bio and Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science, ICT and Future Planning (no. 2012M3A9C6050363).
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
The authors declare no conflict of interest.
Supplementary Material
References
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663–676. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131: 861–872. [DOI] [PubMed] [Google Scholar]
- Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature 2013; 502: 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet 2013; 14: 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro MJ, Kempe H, Verschure PJ. Concise review: the dynamics of induced pluripotency and its behavior captured in gene network motifs. Stem Cells 2013; 31: 838–848. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol 2010; 28: 521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010; 7: 51–63. [DOI] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 2010; 7: 64–77. [DOI] [PubMed] [Google Scholar]
- Xu Y, Wei X, Wang M, Zhang R, Fu Y, Xing M et al. Proliferation rate of somatic cells affects reprogramming efficiency. J Biol Chem 2013; 288: 9767–9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unternaehrer JJ, Zhao R, Kim K, Cesana M, Powers JT, Ratanasirintrawoot S et al. The epithelial-mesenchymal transition factor SNAIL paradoxically enhances reprogramming. Stem Cell Rep. 2014; 3: 691–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 2012; 150: 1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 2012; 151: 1617–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007; 448: 318–324. [DOI] [PubMed] [Google Scholar]
- Lee TH, Song SH, Kim KL, Yi JY, Shin GH, Kim JY et al. Functional recapitulation of smooth muscle cells via induced pluripotent stem cells from human aortic smooth muscle cells. Circ Res 2010; 106: 120–128. [DOI] [PubMed] [Google Scholar]
- Choi HW, Kim JS, Choi S, Jang HJ, Kim MJ, Choi Y et al. Neural stem cells achieve and maintain pluripotency without feeder cells. PLoS ONE 2011; 6: e21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Seng S, Sekine M, Hinton C, Fu Y, Avraham HK et al. Vascular endothelial growth factor mediates intracrine survival in human breast carcinoma cells through internally expressed VEGFR1/FLT1. PLoS Med 2007; 4: 1101–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, van Oosten AL, Castelo-Branco G, Hall J, Smith A, Silva JC. Nanog overcomes reprogramming barriers and induces pluripotency in minimal conditions. Curr Biol 2011; 21: 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods 2011; 8: 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Liu J, Han Q, Qin D, Xu J, Chen Y et al. Towards an optimized culture medium for the generation of mouse induced pluripotent stem cells. J Biol Chem 2010; 285: 31066–31072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Liu J, Chen Y, Yang J, Chen J, Liu H et al. Rational optimization of reprogramming culture conditions for the generation of induced pluripotent stem cells with ultra-high efficiency and fast kinetics. Cell Res 2011; 21: 884–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Xie M, Laurent T, Ding S. Progress in the reprogramming of somatic cells. Circ Res 2013; 112: 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Cañamero M et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 2009; 460: 1136–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol 2008; 26: 795–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Ambasudhan R, Yuan X, Li W, Hilcove S, Abujarour R et al. A chemical platform for improved induction of human iPSCs. Nat Methods 2009; 6: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Beard C, Hanna J, Jaenisch R. Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nat Methods 2010; 7: 56–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanner F, Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development 2010; 137: 3351–3360. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J et al. The ground state of embryonic stem cell self-renewal. Nature 2008; 453: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 2007; 134: 2895–2902. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell 2008; 2: 10–12. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol 2008; 26: 101–106. [DOI] [PubMed] [Google Scholar]
- Shaul YD, Seger R. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim Biophys Acta 2007; 1773: 1213–1226. [DOI] [PubMed] [Google Scholar]
- Romeo Y, Zhang X, Roux PP. Regulation and function of the RSK family of protein kinases. Biochem J 2012; 441: 553–569. [DOI] [PubMed] [Google Scholar]
- Posern G, Treisman R. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol 2006; 16: 588–596. [DOI] [PubMed] [Google Scholar]
- Arsenian S, Weinhold B, Oelgeschläger M, Rüther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J 1998; 17: 6289–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser S, Impey S, Trinh K, Xia Z, Storm DR. SRF-dependent gene expression is required for PI3-kinase-regulated cell proliferation. EMBO J 2000; 19: 4955–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 2013; 502: 65–70. [DOI] [PubMed] [Google Scholar]
- Davis RJ. Transcriptional regulation by MAP kinases. Mol Reprod Dev 1995; 42: 459–467. [DOI] [PubMed] [Google Scholar]
- Sandbo N, Kregel S, Taurin S, Bhorade S, Dulin NO. Critical role of serum response factor in pulmonary myofibroblast differentiation induced by TGF-beta. Am J Respir Cell Mol Biol 2009; 41: 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Vasishtha M, Prywes R. Activation and repression of cellular immediate early genes by serum response factor cofactors. J Biol Chem 2010; 285: 22036–22049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli RR, Segal DJ, Dreier B, Barbas CF 3rd. Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci USA 1998; 95: 14628–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest 2006; 116: 36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J, Sun Y, Singh A, Dalton S. MYC/MAX control ERK signaling and pluripotency by regulation of dual-specificity phosphatases 2 and 7. Genes Dev 2013; 27: 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev 1998; 12: 2048–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Bertone P, Smith A. Identification of the missing pluripotency mediator downstream of leukaemia inhibitory factor. EMBO J 2013; 32: 2561–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 2009; 460: 118–122. [DOI] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD, Whitmarsh AJ. MAP kinase signalling cascades and transcriptional regulation. Gene 2013; 513: 1–13. [DOI] [PubMed] [Google Scholar]
- Liu J, Han Q, Peng T, Peng M, Wei B, Li D et al. The oncogene c-Jun impedes somatic cell reprogramming. Nat Cell Biol 2015; 17: 856–867. [DOI] [PubMed] [Google Scholar]
- Yeo JC, Jiang J, Tan ZY, Yim GR, Ng JH, Göke J et al. Klf2 is an essential factor that sustains ground state pluripotency. Cell Stem Cell 2014; 14: 864–872. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981; 292: 154–156. [DOI] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell 2009; 4: 487–492. [DOI] [PubMed] [Google Scholar]
- Boroviak T, Loos R, Bertone P, Smith A, Nichols J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat Cell Biol 2014; 16: 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.