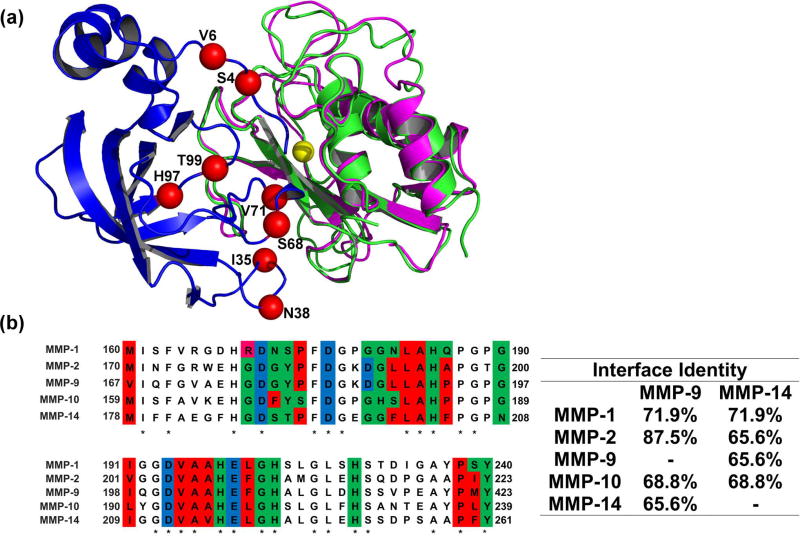
Figure 1.
(A) Structural alignment of MMP-9CAT shown in magenta (PDB: 1L6J) and MMP-14CAT shown in green (PDB: 1BUV) in complex with N-TIMP2 shown in blue. Red spheres indicate positions where mutations were allowed in the designed combinatorial N-TIMP2 library and are labelled with their WT identity. Yellow sphere represents Zn2+ atom in the MMP active site.
(B) Left panel A pairwise alignment of residues on MMP-1CAT, MMP-2CAT, MMP-9CAT, MMP-10CAT, and MMP-14CAT (generated with the aid of EMBL Clustal Omega). Residues located in the WT N-TIMP2:MMP interface are colour coded according to chemical character of their side chains. Red represents hydrophobic residues, green represents hydrophilic residues, pink represents positively charged residues and blue represents negatively charged residues. These interface residues are at a distance of 4Å or less from N-TIMP2. Asterisks indicate that there is a consensus at a particular position. Right panel Percent of amino-acid identity between binding interface residues for various pairs of MMPs is shown.