Figure 4.
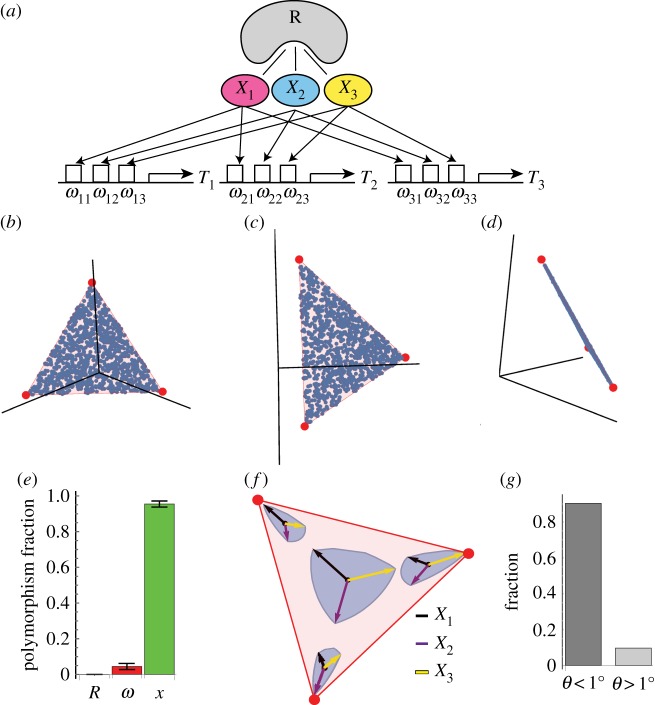
Epistasis due to a molecular mechanism evolves to keep phenotypes within a triangular front (a) A mechanism based on bacterial transcription considers genes whose promoters are regulated by 3 regulators (sigma factors) Xi that compete over a limiting factor R (RNA polymerase). The effect of Xi on the promoter of gene j is ωij. (b–d) Phenotypes (blue dots) reside on the Pareto (red triangle) front after 100 000 generations with this mechanism. Panels show the data from three different views. Each mutation changed one of the parameters Xi, ωij or R by multiplying the parameter by number drawn from a lognormal distribution LN(1,1). (e) Fraction of prevalent polymorphisms (greater than 1% of genomes) in each of the parameters R, ωij (summed over all ωs) and Xi (summed over X1, X2, X3). Median and median deviation over 100 different simulations is shown at 100 000 generations. (f) Effect of mutations that change Xi by 10%. Arrows are 10% changes in X1 (black arrow), X2 (purple arrow) or X3 (yellow arrow), and the blue contour is the boundary of the effects of simultaneously changing X1, X2 and X3 so that the total change is 10%. Mutation effects are magnified by 12 for illustration. Mutations show epistasis, by having different effects at different genetic backgrounds that correspond to four different phenotypes on the front. (g) Fraction of angles with respect to the plane of the triangle that are smaller (left) or bigger (right) than 1°, for all polymorphisms present at greater than 1% of the genomes. Each polymorphism was evaluated in all of the genomes in which it appears, because the angle can vary due to epistasis. Results are based on simulation with parameters N = 1000, μ = 0.05, k = 5, free-recombination. Each mutation changed one of the parameters Xi.