FIGURE 2.
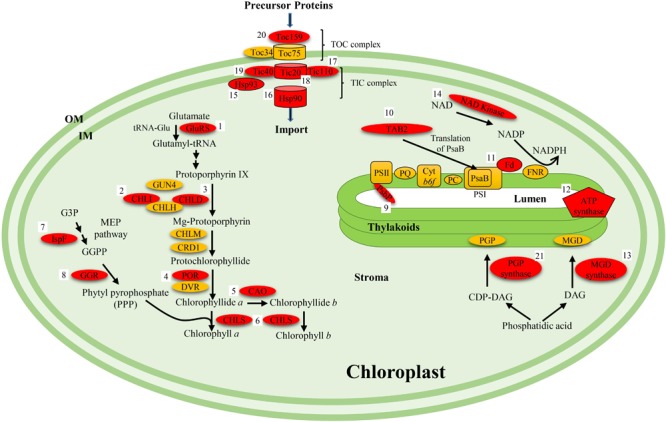
Summary of various proteins involved in yellow foliar phenotypes in plants showing their metabolic functions. The characterized proteins that have been functionally linked with the yellow phenotype are highlighted in red. Numbers 1 through 21 represent names of the mutants identified for the proteins represented in the pathway. (1) cde1 (Os); (2) chl9 (Os), y11 (Gm), and cd5 (Gm); (3) ygl7 (Os), ygl7 (Os), chl1 (Os), and ygl98 (Os); (4) fgl (Os); (5) cao1 (Os); (6) ygl1 (Os); (7) 505ys (Os); (8) lyl1-1 (Os); (9) psbP (Gm); (10) tab2 (Gm); (11) 501ys (Os); (12) yl1 (Os) and ys83 (Os); (13) mgd1 (At); (14) nadk2 (At); (15) hsp93 (At); (16) hsp90c (At); (17) tic110 (Gm); (18) tic20 (At); (19) tic40 (At); (20) toc159 (At); (21) pgp1 (At). Abbreviations in the parenthesis after the mutant name represent the species in which the mutant was identified (At, Arabidopsis thaliana; Os, Oryza sativa; Gm, Glycine max). GluRS, glutamyl-tRNA synthase; GUN4, genomes uncoupled 4; CHLD, CHLH, CHLI are subunits of the Mg-chelatase enzyme; CRD1, copper response defect 1; POR, protochlorophyllide oxidoreductase; DVR, divinyl reductase; CAO, chlorophyll a oxygenase; CHLS, chlorophyll synthase; TICs, translocon at the inner envelope membrane of chloroplast; TOCs, translocon at the outer envelope membrane of chloroplast; PSI, photosystem I; PSII, photosystem II; PQ, plastoquinone; PC, plastocyanin; Cyt b6F, cytochrome b6f; PsbP, component protein of PSII; DAG, diacylglycerol; CDP-DAG, cytidine diphosphate diacylglycerol; PGP, phosphatidyl glycerol phosphate; MGD, monogalactosyl diacylglycerol; TAB2, translation of PsaB 2; PsaB, component protein of PSI; Hsp, heat shock protein; NAD, nicotinamide adenine dinucleotide; FNR, ferredoxin NADP+ reductase; G3P, glycerlaldehyde 3-phosphate; GGPP, geranylgeranyl pyrophosphate; OM, outer membrane; IM, inner membrane.
