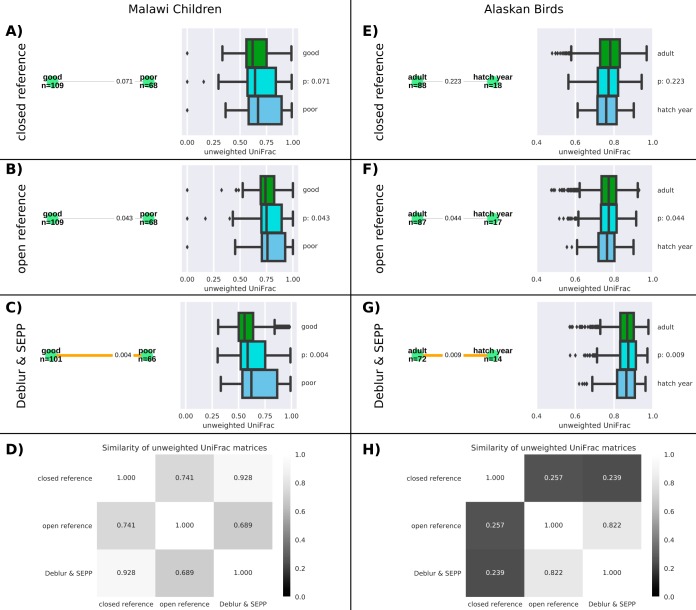
FIG 3 .
Higher sub-OTU resolution, in combination with SEPP phylogenies, exposed relevant ecological signals. (A to D) For the Malawi children, the same 7,554,708 reads from 179 samples (150 nt; mean number of reads per sample, 42,205) were processed by “closed-reference” OTU picking (A), “open-reference” OTU picking against the same reference database (B), and the sub-OTU method “Deblur” (C), and correlation via Mantel tests for unweighted Unifrac beta diversities were computed (D). (E to G) For the Alaska birds, a total of 5,932,450 reads from 137 samples (125 nt; mean number of reads per sample, 43,303) were processed with both methods mentioned above. Pairwise testing between sample groups was performed via PERMANOVA with 9,999 permutations. Statistically significant differences between groups are indicated via bold orange edges, while nonsignificant edges are colored gray. Green boxes at the right side of panels A, B, and C summarize pairwise beta diversity distances within the group of “good” samples, and the dark blue boxes represent distances within “poor” samples. The cyan-colored boxes show between-group distances, i.e., all pairwise distances between “good” and “poor” samples. Similarly, the green, dark blue, and cyan boxes in panels E (closed-reference OTU picking), F (open-reference OTU picking), and G (Deblur) summarize pairwise distances within “adult” and “hatch year” data and between samples, respectively, and correlation via Mantel tests for unweighted Unifrac beta diversities were computed (H).