Figure 2.
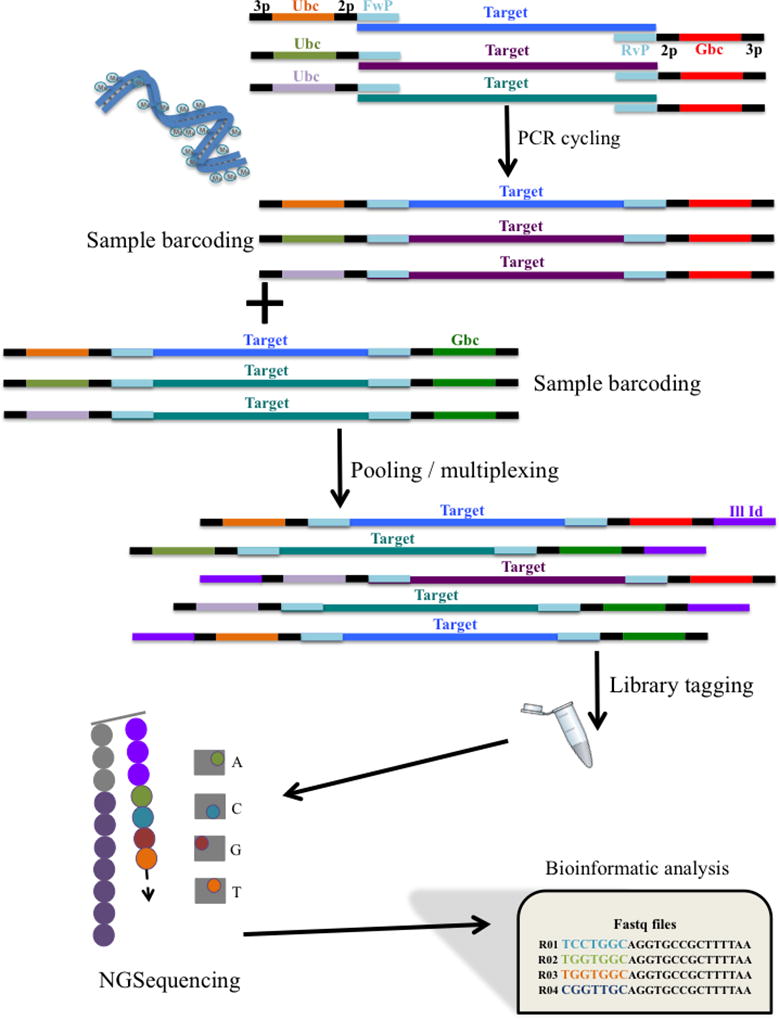
Simplified workflow for HPV typing using NGS methods. During PCR each sample is uniquely barcoded via the primer pair. The use of multiple barcodes allows for larger number of samples to be tested. NGS has its own chemistry that recognizes the presence of an Illumina Index (Ill Id) for sequencing purposes. During sequencing in the flow cell, as each nucleotide is incorporated releasing a flash of light, a picture is retained and analyzed by specialized software to identify the specific nucleotide incorporated. Raw data files comprise million of sequence reads from a pool of samples that need to be analyzed using bioinformatic tools (3p: 3bp-pad; Ubc: unique forward barcode sequence; 2p: 2bp-pad; FwP: forward primer sequence; HPV target: HPV amplicon of interest; RvP: reverse primer sequence; Gbc: general reverse barcode sequence).
