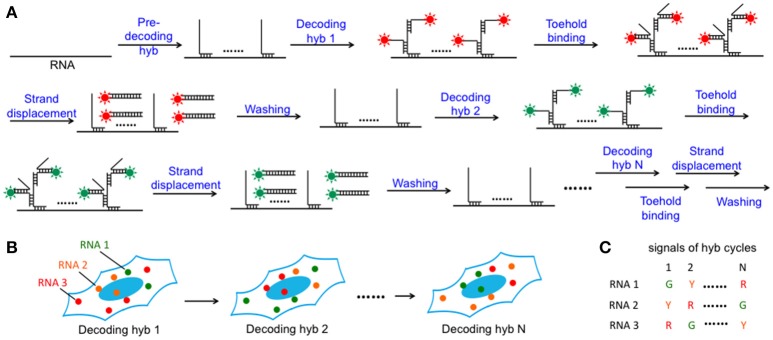
Figure 1.
Highly multiplexed single-cell in situ RNA analysis with SFO. (A) Each transcript is first hybridized with a set of pre-decoding probes, which have varied target-binding sequences to hybridize to the different regions on the target RNA and the shared decoding sequence to recruit SFO as decoding probes. After imaging, the hybridized SFO is removed by strand displacement reactions. Through reiterative cycles of SFO hybridization, fluorescence imaging and strand displacement, the target RNA is sequentially stained by a set of SFO labeled with varied fluorophores. (B) Schematic diagram of the N cycles of hybridization images. In each cycle, individual transcript is visualized as a single spot with a specific color. (C) As RNA molecules remain in place during different hybridization cycles, different RNA species can be identified by the unique color sequences.