Fig. 1.
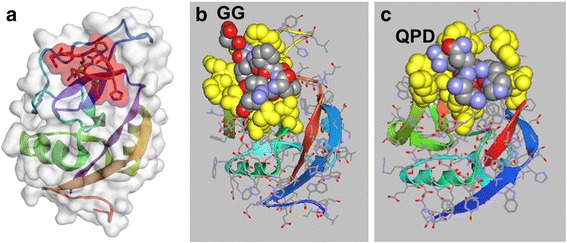
Docking of sv6D to receptors. a In silico docking of an arm of sv6D (NQHTPRGG) to the carbohydrate-recognition domain of ASGPR-1 (accession number 1DV8) with CABS-dock (RMSD = 0.7611 Å) [60]. The peptide is enclosed in red shading that delineates the space-filling molecular structure. b The structure in (a) as rendered in ArgusLab 4.0.1 (predicted binding energy, ΔG’ = − 40 kJ/mol). Amino acids in the binding site that interact with the peptide are colored in yellow as space-filling structures. sv6D is colored as carbon, grey; nitrogen, blue; and oxygen, red. A portion of the linker to the remainder of the tetravalent structure is indicated as GG. c The structure of CLEC10A was generated with SWISS-MODEL Deep View [58, 59] from the structure of ASPGR-1. Docking was modeled with CABS-dock (RMSD = 1.421 Å) and downloaded into ArgusLab 4.0.1 (predicted binding energy, ΔG’ = − 38 kJ/mol). The binding site and peptide are colored as in (b). The location of the QPD sequence is indicated. Helical and beta-strand secondary structures of the protein are shown as colored ribbons
