Abstract
Cerebellar Abiotrophy (CA) is a neurodegenerative disease in Arabian horses affecting the cerebellum, more specifically the Purkinje neurons. Although CA occurs in several domestic species, CA in Arabian horses is unique in that a single nucleotide polymorphism (SNP) has been associated with the disease. Total RNA sequencing (RNA-seq) was performed on CA-affected horses to address the molecular mechanism underlying the disease. This research expands upon the RNA-seq work by measuring the impact of the CA-associated SNP on the candidate gene MutY homolog (MUTYH) and its regulation, isoform-specific expression and protein localization. We hypothesized that the CA-associated SNP compromises the promoter region of MUTYH, leading to differential expression of its isoforms. Our research demonstrates that the CA-associated SNP introduces a new binding site for a novel transcription factor (Myelin Transcription Factor-1 Like protein, MYT1L). In addition, CA-affected horses show differential expression of a specific isoform of MUTYH as well as different localization in the Purkinje and granular neurons of the cerebellum.
Keywords: MUTYH, Horse, Cerebellar abiotrophy
1. Introduction
Cerebellar Abiotrophy (CA) is a neurodegenerative disease affecting the cerebellum and is likely caused by an intrinsic metabolic disorder (Siso et al., 2006). There are incidences of CA in multiple species (de Lahunta, 1990), including humans. Although there is a common mode of inheritance, there is no candidate gene consistently affected across species. In the horse, CA affects the Arabian breed and manifests itself between days to months after birth, resulting in apoptosis of Purkinje neurons and secondary loss of granular neurons (Blanco et al., 2006). CA presents as an ataxic phenotype including uncoordinated limb movement, head tremors, lack of menace response and difficulty rising (DeBowes et al., 1987). CA in the horse is associated with a single nucleotide polymorphism (SNP), which is inherited in an autosomal recessive manner (Brault et al., 2011). The CA-associated SNP is located in the coding region of the target of Egr1 gene (TOE1). However, the gene architecture surrounding this SNP also places it in the promoter region of MutY homolog (MUTYH) (Scott et al., 2017). Thus, MUTYH was further characterized in the cerebella of control and CA affected horses.
MUTYH is a DNA glycosylase that repairs oxidative lesions in DNA. It functions on both nuclear and mitochondrial DNA to specifically remove adenines that are mis-paired to the oxidative lesion, 8-oxoguanine (Ohtsubo et al., 2000). MUTYH also can induce apoptosis under conditions of high oxidative stress in the nucleus or mitochondria (Oka and Nakabeppu, 2011). Expression of MUTYH is relatively high in the central nervous system (CNS) and detectable in the cerebellum of mice (Ichinoe et al., 2004) and rats (Englander et al., 2002). Multiple isoforms of MUTYH are found in the mouse (Ichinoe et al., 2004), rat (Englander et al., 2002) and human (Plotz et al., 2012), with differential localization of specific isoforms to the nucleus or to mitochondria. In the CNS, where most of the cells are post-mitotic, the majority of MUTYH expression is from the mitochondrial isoforms (Plotz et al., 2012). Although TOE1 appears to be a likely candidate gene, compromised MUTYH gene regulation is more satisfactory in explaining the cerebellum-specific neurodegeneration. Expression of TOE1 on the gene and isoform level, as indicated by our previous RNA-seq data, has also been found to be unchanged between CA-affected and controls horses (Scott et al., 2017). The variability created through MUTYH and its response to oxidative damage also permits the flexibility essential in explaining the variable onset and severity of the CA phenotype.
Although first known for its association with MUTYH-associated polyposis, a syndrome with increased risk of colorectal cancer (Al-Tassan et al., 2002); MUTYH has also been implicated in multiple neurodegenerative diseases. In a Friedreich’s ataxia mouse model, there was an increase in 8-oxoguanine and MUTYH in the microglia within the cerebellum (Shen et al., 2016). It was proposed that MUTYH and PARP-1 participated in microglial activation in response to DNA damage in the cerebellum (Shen et al., 2016). Individuals with Parkinson’s disease exhibit an increase in mitochondrial MUTYH expression within the substantia nigra (Arai et al., 2006). Regarding CA in the horse, total MUTYH expression was found to be unchanged in horses affected by CA (Scott et al., 2017). However, with the development of a new equine transcriptome (Mansour et al., 2016) and RNA-seq work done in CA-affected horses (Scott et al., 2017); additional isoforms of MUTYH have been identified, allowing for a more in-depth analysis of MUTYH isoform usage. We hypothesize that the CA-associated SNP compromises the promoter region of a MUTYH isoform, causing differential expression of a subset of the MUTYH isoforms. Here we report the complete characterization the MUTYH promoter region, quantification of MUTYH isoform expression levels, and immunostaining to identify MUTYH localization in CA-affected cerebella.
2. Results
2.1. MUTYH promoter methylation and protein binding in CA-affected horses
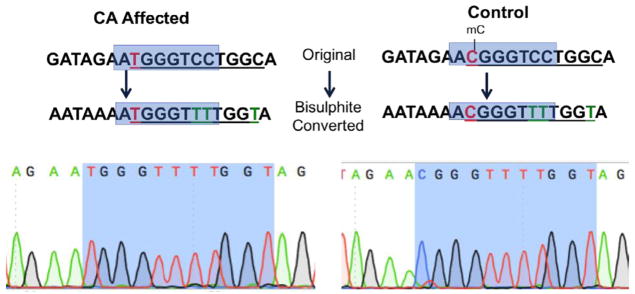
Bisulphite sequencing revealed that the CA-associated G>A SNP affects the local methylation pattern. The wildtype site is methylated in control horses, but the affected horses showed no methylation due to the alteration from a CG site to a TG site (Fig. 1). Fourteen horses were analyzed, all details of these horses can be found in Supplementary Table 1.
Fig. 1.
Differential promoter behavior in CA-affected horses versus control horses. The CA-associated SNP (in red) changes a wild type methylated CG site into a non-methylated TG site, mC denotes a methyl group on a cytosine, green T represents cytosines that are not methylated and thus converted during bisulphite conversion and the blue box in the text version of the sequence represents the highlighted blue area in the electropherogram. This is a representative example from a 1-year-old control versus 9-month-old CA-affected horse.
A pull down of a 50 bp probe centered over the position of the CA-associated SNP from the promoter region of MUTYH shows a consistent, unique binding of Myelin Transcription Factor-1 Like protein (MYT1L) exclusively on the MUTYH promoter oligo containing the CA-associated SNP (Table 1). Additionally, TUBA1B was found to bind the oligo containing the CA-associated SNP, but not the wild-type sequence, however, it was present in the control wash as well. Mass spectrometry detected the binding of Myelin Basic Protein (MBP) to both the wild-type and CA-associated SNP-containing oligos (Supplementary Fig. 1, Table 1). Mass spectrometry was also performed on the washes to elucidate proteins that may be weakly associated with MUTYH promoter region and, although the washes between the control and CA-affected promoter are quite similar, the CA-affected wash also uniquely contained the protein ALDOC, also known as zebrin II. All mass spectrometry results and protein identification percentages can be found in Supplementary Table 2.
Table 1.
Proteins identified using mass spectrometry with a peptide FDR threshold of 5% and protein identification threshold of 100%, as calculated by Scaffold v4.4.6.
| Sample | Proteins identified with Mass Spec, size of protein (kDa) |
|---|---|
| Control, wash 1 | MBP (19), HBB (16), HBA (15), TUBB4A (48), TUBA1B (50) |
| Control, pulldown | MBP (19) |
| CA-affected, wash 1 | HBB (16), HBA (15), TUBB4A (48), TUBA1B (50), ALDOC (39), F7C1K6-uncharacterized proteins (28), CKB (43), GAPDH (36) |
| CA-affected, pulldown | MBP (19), TUBA1B (50), MYT1L (133) |
2.2. Bias in MUTYH isoform usage in CA-affected horses
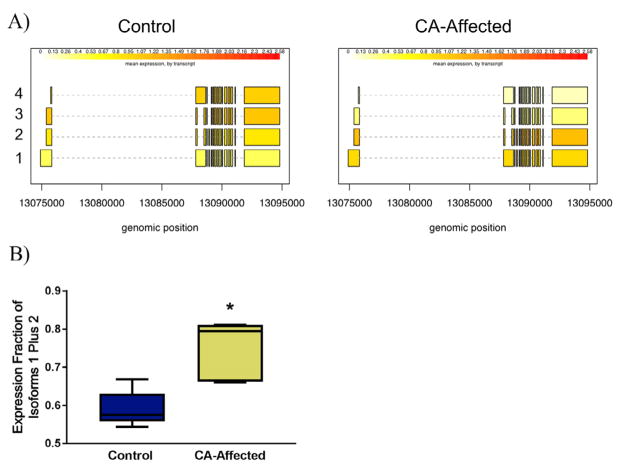
Transcriptomic analysis of MUTYH usage based on RNA-seq data reveals differential isoform usage across all tissues (data not shown), and more specifically differential MUTYH isoform usage in the cerebellum between control and CA-affected horses, with isoforms 1 plus 2 being favored in cerebella from CA-affected horses (Fig. 2A), n = 12, full details of horses can be found in Supplementary Table 1. The ddPCR analysis confirmed that the majority of MUTYH expression in the cerebellum originates from MUTYH isoforms 1 plus 2 in CA-affected horses compared to healthy control cerebella (p = .03, Mann-Whitney U test, two-tailed) (Fig. 2B). While primers could only be designed to determine expression of MUTYH isoforms 1 plus 2, RNA-seq data demonstrated that the control horses bias MUTYH isoform 3 and 4 expression (Fig. 2A).
Fig. 2.
Bias in expression of MUTYH isoforms 1 and 2 in CA-affected horses. RNA-seq data initially characterized this bias (isoforms are labeled with Arabic numerals on the left, expression units are fragments per kilobase per million of reads, fpkm) (A). Using ddPCR, a significant increase in expression of MUTYH isoform 1 and 2 was seen in CA-affected horses (p = .0317, Mann-Whitney U test, two-tailed, n = 15) (B).
2.3. Localization patterns of MUTYH in CA-Affected horses
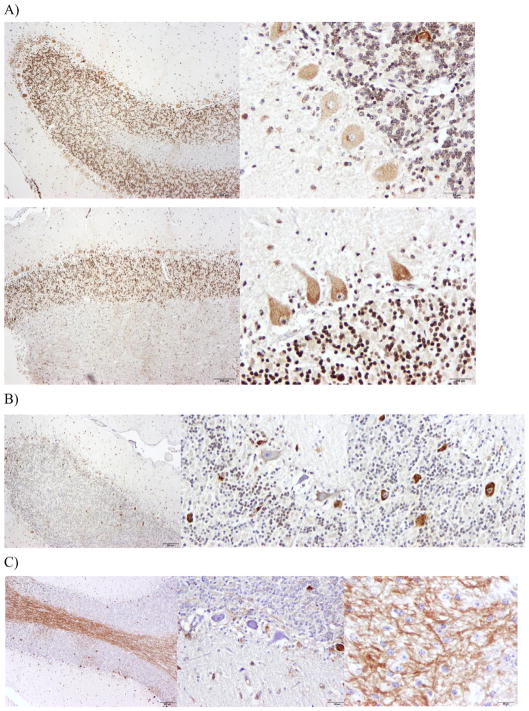
In control horses MUTYH protein is localized to punctae in the cytoplasmic region of Purkinje neurons, strongly to the nuclei of granular neurons and to interspersed nuclei of interneurons in the molecular layer (Fig. 3A), n = 2. In CA-affected horses MUTYH protein is localized to the cytoplasmic region of Purkinje neurons, often too intensely to distinguish punctae, and to nuclei of interneurons. However, there are two differential patterns of MUTYH protein localization in CA-affected horses (Fig. 3B & C), n = 8. In some individuals, mainly the older, less severely affected horses, MUTYH localizes more strongly to the superficial layer of granular neurons, closer to the Purkinje neuron layer, and is heavily expressed in Purkinje neurons that are ectopically located in the granular layer (Fig. 3B). In another group of CA-affected horses, the younger and more severely affected horses, there is an intense staining of the white matter, presumably in the axons of the Purkinje neurons projecting into the adjacent white matter, with little staining of granular neurons and selective staining of Purkinje neurons (Fig. 3C). We are unable to distinguish whether there is MUTYH staining in the astrocytes or microglia, further staining with co-markers for these cell types would need to be done to discern this.
Fig. 3.
Differential localization of MUTYH in control (A) and CA-affected horses (B, C). Using the primary antibody rabbit anti-MUTYH (LS-C354036, LifeSpan Biosciences, Inc) and the secondary biotinylated goat anti-rabbit IgG, with DAB staining for visualization. Healthy horses show cytoplasmic staining in Purkinje neurons and nuclear staining in granular neurons for MUTYH (A). CA-affected horses show two patterns of localization for MUTYH. One pattern is cytoplasmic staining in Purkinje neurons and nuclear staining for superficial layer of granular neurons for MUTYH (B). The other pattern is random cytoplasmic staining in Purkinje neurons, no staining in the granular neurons and intense staining of the white matter (C).
We also observed a case of a 19-year-old horse (mare) considered asymptomatic throughout life as it presented no sign of behavioral and ataxic traits typical of the disease, despite being homozygous for the CA-associated SNP. This horse had unique MUTYH patterning differing from controls and similar to a milder CA histological phenotype (Supplementary Fig. 2). As this is, to our knowledge, the first and only asymptomatic horse characterized on a molecular and histological level, no conclusion can be drawn regarding this phenotype known to occur in some horses homozygous for CA-associated SNP. Nevertheless, the information is useful to have as similar cases may be investigated in the future.
3. Discussion
Cell and subsequent tissue-specific identity is based upon differential usage of the genome. Avenues to generate cell and tissue-specific gene expression begin with modifying the accessibility of genomic DNA in a tissue-specific manner, such as the methylation process. Next, there is the balance between promoter complexity along with temporal and tissue-specific expression of transcription factors complementing the promoter. Finally, there can be tissue-specific usage of various isoforms of a gene. MUTYH has shown differences in its promoter, isoform expression, and localization in the cerebella of CA-affected horses. Crucially, MUTYH gene expression coincides with the tissue specificity, as well as the spectrum of onset and severity of the CA phenotype.
Methylation patterns in promoters have been associated with transcriptional silencing. Although exact mechanisms have not been established, observation of increased uniform methylation negatively correlating with chromatin accessibility (Siegfried et al., 1999) and occupancy of transcription factor binding sites, by methyl groups, leading to reduced transcription factor expression (Thurman et al., 2012) have been proposed as potential mechanisms. The CA-associated SNP not only results in a G>A transversion, but it also results in the loss of methylated CpG site in the MUTYH promoter. With this loss of methylation, there was unique binding of the transcription factor MYT1L only to the MUTYH promoter containing the CA-associated SNP. MYT1L is a member of the a highly conserved superfamily of zinc finger transcription factors (Kim and Hudson, 1992), most commonly recognized for its role in transcriptional repression leading to neuronal stem cell reprogramming (Mall et al., 2017). Although the previously defined binding sequence associated with MYT1L; AAGTT, was not found in either the CA-associated SNP containing or wildtype MUTYH promoter, this motif is found in only 30% of MYT1L targets (Mall et al., 2017). Additionally, the AAGTT motif was more commonly found in MYT1L targets that were downregulated versus upregulated (Mall et al., 2017). Whether MYT1L increases or decreases the expression of certain isoforms of MUTYH has yet to be determined. Further work characterizing MYT1L binding to the MUTYH promoter is required.
Within MUTYH expression, there is evidence of tissue-specific isoforms, as well as environment dependent isoforms. In the rat, Englander et al. (2002) found three isoforms of MUTYH specific to the brain, of which some are inducible in the presence of 8-oxoguanine. Brain-specific MUTYH isoforms have also been found in the mouse (Ichinoe et al., 2004), with some indications of brain-specific isoforms in the horse. While we previously postulated that total MUTYH expression was down-regulated (Brault et al., 2011) or unchanged (Scott et al., 2017) in CA-affected horses, the identification of specific isoforms has allowed us to determine that MUTYH isoforms are differentially expressed in the equine cerebellum, with isoforms 1 plus 2 (isoforms containing exon 3) having significantly increased expression in the cerebella from CA-affected animals. Although further work must be done validating isoforms 3 and 4 expression, it appears to be biased in healthy horses. In humans, over 15 isoforms of MUTYH have been identified, with presumably different regulatory mechanisms, as suggested by the three alternate first exons (Plotz et al., 2012). The different isoforms often differ in their first three exons, with the major differences amounting to localization in the nucleus or the mitochondria (Plotz et al., 2012; Oka et al., 2014). These isoforms display a temporal pattern of expression with nuclear isoforms often being expressed early in life, with a switch to mitochondrial isoforms later (Lee et al., 2004). Presumably, the usage of different promoters dictating the specific MUTYH isoform expression is what permits this temporal and spatial restraint. Therefore, a change in the promoter region of MUTYH may have the largest impacts on a specific tissue during a narrow time frame, as seen in equine CA. Although a specific MUTYH isoform localizing to the cerebellum has not been identified, perhaps multiple combinations of temporal and spatial expression during cerebellum development account for the variable onset and severity of phenotype seen in equine CA.
MUTYH localization in a healthy equine cerebellum is similar to that documented in the mouse and rat. Within the rat cerebellum, MUTYH is predominantly in the Purkinje neurons, moderately present in the granular layer and shows no expression in astrocytes (Englander et al., 2002). In the mouse, MUTYH is present in the granular, Purkinje and molecular layer of the cerebellar cortex (McCarthy, 2006). Interestingly in the horse, it seems as though the nuclear isoform of MUTYH is present in the granular neurons, whereas the Purkinje neurons show a speckled cytoplasmic stain, perhaps indicative of mitochondria-localized MUTYH protein. Typically, post-mitotic cells predominantly express the mitochondrial isoform (Lee et al., 2004); therefore it is unclear why the granular neurons would harbor the nuclear MUTYH isoform. Despite what seems to be a relatively conserved distribution of MUTYH protein across species, the CA-affected horses deviate from this distribution showing two different patterns of MUTYH localization. In the younger, more phenotypically severe CA-affected horses, there was intense staining of MUTYH protein in the white matter, along with a lack of granular neuron nuclei staining for MUTYH. This could strengthen the assumption that the expressed MUTYH protein is localizing to the mitochondria, especially the mitochondria in the Purkinje neuron axons in CA affected horses. Instrumental in dysfunctional mitochondria, oxidative damage has been found to increase the size and number of motile mitochondria in Purkinje neuron axons (Errea et al., 2015). Purkinje neurons have also been speculated to be less tolerant of mitochondrial dysfunction during development due to occlusion of the extensive branch points in their elaborate dendrites by these mitochondria (Chen et al., 2007). The second pattern of MUTYH protein localization seen in CA-affected horses included staining of the subset of granular neurons closest to the Purkinje neuron layer and the strong presence of MUTYH in the ectopic Purkinje neurons. This selective granular neuron staining for MUTYH may reflect granular neuron migration under the neurodegenerative conditions of CA. There are different rates of migration depending on factors such as proliferation of granular neuron precursors in the external granular layer (Wechsler-Reya and Scott, 1999) or the synaptic contacts made early on by granular neurons. Granular neurons forming early synapses with zebrin I-negative Purkinje neurons have been speculated to migrate faster into the inner granular layer and establish unique connections with the Purkinje neurons (Consalez and Hawkes, 2012). Characterizing the granular neurons retaining nuclear MUTYH protein staining could be useful in elucidating the temporal component of when the CA-associated SNP is most detrimental to Purkinje neurons. It is unclear whether the different MUTYH staining patterns seen in CA-affected individuals is due to age or severity of CA phenotype and thus further work is required. Importantly, the different localization patterns of MUTYH seen in CA-affected horses compared to healthy controls suggests that MUTYH does have a role in the CA disease phenotype.
Our research has demonstrated that MUTYH is a candidate gene affected by the CA-associated SNP on regulatory, gene expression and protein localization levels. Further work to characterize the remaining Purkinje neurons and their mitochondrial health would help resolve the pattern and mechanism by which MUTYH affects Purkinje neurons. Particular emphasis on zebrin II expression and how it relates to Purkinje neuron degeneration in equine CA is the most likely next step to understanding the patterns underlying CA phenotype. Strengthening the relationships between MUTYH expression, localization and cerebellar specificity would also be beneficial in understanding the explicit degeneration of the cerebellum and the variation in phenotype observed within the cerebellar cortex of CA-affected horses. This research has identified means by which MUTYH expression could be influenced by the CA-associated SNP in a tissue-specific manner. Further work characterizing MUTYH in relation to the behavioral phenotype and other cerebellar molecular markers in CA-affected horses would be beneficial in understanding the understated molecular heterogeneity present in the cerebellar cortex and variability in the behavioral phenotype.
4. Methods
4.1. Bisulphite sequencing
Genomic DNA (gDNA) from six controls and six CA-affected cerebella (cerebellar cortex) were extracted using Quick-DNA Universal kit (Zymo Research, Irvine, CA, USA), according to manufacturer’s protocol. All samples, along with age, sex and which analyses were performed on them can be found in Supplementary Table 1. Approximately 500 ng of gDNA from each sample was bisulphite converted using the EZ DNA Methylation kit (Zymo Research). The candidate 100 bp promoter region of MUTYH had primers forward: GGAGAATATATTAAGATTTGGAAGGAGA and reverse: AAATACTTCCTAAAACTAACTCCCAACC. The polymerase chain reaction (PCR) was then performed on 160 ng of bisulphite converted DNA. PCR amplification cycling conditions consisted of a denaturing temperature of 95 °C for 2 min, followed by 40 cycles of 95 °C for 30 s, 58 °C for 45 s and 72 °C for 30 s, and a final extension of 72 °C for 5 min. The PCR reactions contained 5X green GoTaq® Reaction Buffer, 0.2 mM of each dNTP, 2.5 mM MgCl2, 0.5 uM each primer and 1.25 U of GoTaq® DNA Polymerase (Promega, Madison, WI, USA). PCR products were then gel extracted using the QIAquick Gel extraction kit (Qiagen, Hilden, Germany) and sent for Sanger sequencing (Quintara Biosciences, San Francisco, CA, USA).
4.2. Extraction of nuclear proteins from equine cerebellum, MUTYH promoter pull down and mass spectrometry
Whole cerebellum from a healthy five-year-old thoroughbred mare was harvested and immediately snap-frozen in liquid nitrogen. Forty mg of cerebellar cortex was then cut and nuclear proteins were isolated using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (ThermoScientific, Waltham, MA, USA) according to manufacturer’s instructions. The promoter pull down protocol was adapted from Carey et al. (2010). Briefly, two biotinylated oligo probes representing a 50 bp seq of the MUTYH promoter with and without the CA-associated SNP in the middle (AAGCAGGC GAGGCCCAGGGAGAGGATAGAACGGGTCCTGGCAGCATGACA) were incubated and bound to streptavidin beads (Pure Biotech LLC, Middlesex, NJ, USA) using a rotator (40 rpm) for one hour at room temperature. These beads were then incubated with cerebellar nuclear extract on a rotator (40 rpm) for 30 min. Beads were washed twice and oligos along with bound proteins were resuspended in TEN buffer and removed by boiling for 5 min. Half of the resulting extracts (20ul) were run on a SDS-PAGE gel, which was then stained with Coomasie blue reagent. The other half was sent for mass spectrometry (Campus Mass Spectrometry Facilities, University of California-Davis) and analyzed using Xevo G2 QTof coupled to a nanoAcquity UPLC system (Waters, Milford, MA). Mass spectrometry data were recorded for 60 min for each run and controlled by MassLynx 4.1 (Waters, Milford, MA). Acquisition was set to positive polarity under resolution mode. Mass range was set from 50 to 2000 Da. Capillary voltage was 3.5 kV, sampling cone at 25 V, and extraction cone at 2.5 V.
4.3. Mass spectrometry data analysis
RAW MSe files were processed using Protein Lynx Global Server (PLGS) version 2.5.3 (Waters, Milford, MA). Processing parameters consisted of a low energy threshold set at 200.0 counts, an elevated energy threshold set at 25.0 counts, and an intensity threshold set at 1500 counts. The UniProt (www.uniprot.org) equine databank was used for peak identification. Searches were performed with trypsin specificity and allowed for three missed cleavages. Possible structure modifications included for consideration were methionine oxidation and carbamidomethylation of cysteine. For viewing, PLGS search results were exported in Scaffold v4.4.6 (Proteome Software Inc., Portland, OR). Peptide identifications were accepted if they could be established at greater than 5.0% probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 99.9% probability and contained at least 1 identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003).
4.4. RNA extraction, cDNA synthesis and droplet digital PCR
Whole cerebellum cortex samples were collected from nine CA-affected and six control horses and stored in RNAlater® (AM7020, Life technologies) for up to one week at 4 °C and then transferred to −80 °C. RNA was extracted using 1 ml of TRIzol (15596-018, Invitrogen) per 100 mg of tissue and an electric homogenizer (TissueMiser, Fisher Scientific, Pittsburg, PA, USA) was used for mechanical shearing. Chloroform was used for phase separation and RNA was concentrated, eluted, and treated with DNase (Promega, Madison, WI, USA) on Zymo RNA extraction columns (Zymo Research, Irvine, CA, USA) according to manufacturer’s instructions. RNA was resuspended in DEPC-treated water and quantified using a NanoDrop 2000 spectrophotometer (ThermoScientific). cDNA was made with 1 μg of RNA using 200 units of SuperScriptII Reverse Transcriptase (18064, Invitrogen), 0.5 μg of Oligo (dT) 15 (Promega), 10 mM each dNTP mix (RR02AG, TaKara), 5X first strand buffer (Invitrogen), 0.01 MDTT (Invitrogen) and 40 units of RNaseOUT (Invitrogen). RNA, dNTPs, and oligo (dT) 15 were incubated for 5 min at 65 °C then 5x first strand buffer, DTT and RNaseOUT were added and incubated for 1 min at 42 °C. SuperScriptII Reverse Transcriptase was then added and contents were incubated for 50 min at 42 °C and then 70 °C for 15 min. cDNA samples were stored in at −80 °C. ddPCR was performed on the samples mentioned above. One common set of Taqman primers was used with one probe for total MUTYH and one probe for the isoform containing exon 3, isoforms 1 plus 2 (Supplementary Table 2). A QX200™ ddPCR system (Bio-Rad) was used according to the manufacturer’s instructions. The ddPCR reaction mixture (25 μl) contained ddPCR supermix for probes (no dUTP) (catalog number: 186-3023, Bio-Rad), 1.1 μl of each primer/probe mixture (10 μM of each primer and 5 μM of probe) and 100 ng of template cDNA. Droplets were generated using the Droplet Generator with 70 μl of droplet generation oil and 20 μl of above reaction per well. Droplets containing reactions were then cycled through 94 °C for 2 min, followed by 40 cycles of 94 °C for 20 s and 58 °C for 30 s, one cycle of 72 °C for 5 min and held at 4 °C. After completion, the droplets were read using the Droplet Reader and QuantaSoft software was used to analyze the data.
4.5. Immunohistochemistry
Equine cerebella were fixed in 10% neutral buffered formalin, embedded in paraffin, and sectioned (5um). Sections were subsequently rehydrated through incubations in xylene and gradations of ethanol to double distilled water. Heat induced epitope retrieval was performed using Dako target retrieval solution, pH = 6.0 (Dako, Carpinteria, CA, USA) in a vegetable steamer for 18 min at 95 °C. Endogenous peroxidase was quenched using 0.3% hydrogen peroxidase in 70% methanol, and non-specific binding was blocked using 5% normal goat serum in 1X TBS-T. Avidin then biotin blocks were then applied for 15 min each (ThermoFisher Scientific, Grand Island, NY, USA). Sections were incubated overnight at 4 °C with rabbit anti-MUTYH (LS-C354036, LifeSpan Biosciences, Inc) diluted 1:100 in Dako Antibody Diluent (Dako). After washing slides in TBS-T, they were incubated with a secondary biotinylated goat anti-rabbit IgG, diluted 1:400, and a streptavidin-HRP conjugate, diluted 1:200, for 30 min. each. Slides were developed for 3.5 min with the peroxidase substrate kit ImmPACT™ DAB (Vector, Burlingham, CA, USA). Lastly, slides were counterstained with Modified Mayer’s hematoxylin (ThermoFisher Scientific) for 45 s and then rinsed with VWR Bluing agent RTU (VWR, Richmond, IL, USA) for 1 min. Images were observed an Olympus Bx-43 microscope and captured with an Olympus DP-72 camera using DP2-BSW software (Olympus).
Supplementary Material
Acknowledgments
Funding
This project was funded in part by the Arabian Horse Foundation as well as the Animal Science Department and Veterinary Genetics Laboratory, University of California, Davis, USA.
Samples were provided by private donors and University of Pennsylavania, Colorado State University and University of Kentucky. Additional acknowledgement to Dr. Rob Grahn for help with the ddPCR and Dr. Armann Andaya at the UC Davis Campus Mass Spectrometry Facility who was tremendous help with the Mass spectrometry analysis.
Abbreviations
- CA
cerebellar abiotrophy
- SNP
single nucleotide polymorphism
- RNA-seq
total RNA sequencing
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.brainres.2017.10.034.
Footnotes
Author contributions
E.Y.S. wrote the manuscript and conducted the lab work. K.D.W. mentored the immunohistochemistry work and edited the manuscript. J.D.M., C.J.F. and M.C.T. Penedo helped with experimental design, manuscript editing and overall mentorship.
Competing interests
Authors declare no competing interest.
References
- Siso S, Hanzlicek D, Fluehmann G, Kathmann I, Tomek A, Papa V, et al. Neurodegenerative diseases in domestic animals: a comparative review. Vet J. 2006;171(1):20–38. doi: 10.1016/j.tvjl.2004.08.015. [DOI] [PubMed] [Google Scholar]
- de Lahunta A. Abiotrophy in domestic animals: a review. Can J Vet Res. 1990;54(1):65–76. [PMC free article] [PubMed] [Google Scholar]
- Blanco A, Moyano R, Vivo J, Flores-Acuna R, Molina A, Blanco C, et al. Purkinje cell apoptosis in Arabian horses with cerebellar abiotrophy. J Vet Med A Physiol Pathol Clin Med. 2006;53(6):286–287. doi: 10.1111/j.1439-0442.2006.00836.x. [DOI] [PubMed] [Google Scholar]
- DeBowes RM, Leipold HW, Turner-Beatty M. Cerebellar abiotrophy. Vet Clin North Am Equine Pract. 1987;3(2):345–352. doi: 10.1016/s0749-0739(17)30677-6. [DOI] [PubMed] [Google Scholar]
- Brault LS, Cooper CA, Famula TR, Murray JD, Penedo MC. Mapping of equine cerebellar abiotrophy to ECA2 and identification of a potential causative mutation affecting expression of MUTYH. Genomics. 2011;97(2):121–129. doi: 10.1016/j.ygeno.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Scott EY, Penedo MC, Murray JD, Finno CJ. Defining trends in global gene expression in Arabian horses with cerebellar abiotrophy. Cerebellum. 2017;16(2):462–472. doi: 10.1007/s12311-016-0823-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo T, Nishioka K, Imaiso Y, Iwai S, Shimokawa H, Oda H, et al. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28(6):1355–1364. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka S, Nakabeppu Y. DNA glycosylase encoded by MUTYH functions as a molecular switch for programmed cell death under oxidative stress to suppress tumorigenesis. Cancer Sci. 2011;102(4):677–682. doi: 10.1111/j.1349-7006.2011.01869.x. [DOI] [PubMed] [Google Scholar]
- Ichinoe A, Behmanesh M, Tominaga Y, Ushijima Y, Hirano S, Sakai Y, et al. Identification and characterization of two forms of mouse MUTYH proteins encoded by alternatively spliced transcripts. Nucleic Acids Res. 2004;32(2):477–487. doi: 10.1093/nar/gkh214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander EW, Hu Z, Sharma A, Lee HM, Wu ZH, Greeley GH. Rat MYH, a glycosylase for repair of oxidatively damaged DNA, has brain-specific isoforms that localize to neuronal mitochondria. J Neurochem. 2002;83(6):1471–1480. doi: 10.1046/j.1471-4159.2002.01259.x. [DOI] [PubMed] [Google Scholar]
- Plotz G, Casper M, Raedle J, Hinrichsen I, Heckel V, Brieger A, et al. MUTYH gene expression and alternative splicing in controls and polyposis patients. Hum Mutat. 2012;33(7):1067–1074. doi: 10.1002/humu.22059. [DOI] [PubMed] [Google Scholar]
- Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, et al. Inherited variants of MYH associated with somatic G:C→T: A mutations in colorectal tumors. Nat Genet. 2002;30(2):227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- Shen Y, McMackin MZ, Shan Y, Raetz A, David S, Cortopassi G. Frataxin deficiency promotes excess microglial DNA damage and inflammation that is rescued by PJ34. PLoS One. 2016;11(3):e0151026. doi: 10.1371/journal.pone.0151026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Fukae J, Hatano T, Kubo S, Ohtsubo T, Nakabeppu Y, et al. Upregulation of hMUTYH, a DNA repair enzyme, in the mitochondria of substantia nigra in Parkinson’s disease. Acta Neuropathol. 2006;112(2):139–145. doi: 10.1007/s00401-006-0081-9. [DOI] [PubMed] [Google Scholar]
- Mansour T, Scott EY, Finno CJ, Bellone R, Mienaltowski MJ, Ross PJ, Valberg SJ, Penedo MCT, Murray JD, Brown CT. Tissue resolved, gene structure refined equine transcriptome. BMC Genomics. 2016 doi: 10.1186/s12864-016-3451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried Z, Eden S, Mendelsohn M, Feng X, Tsuberi BZ, Cedar H. DNA methylation represses transcription in vivo. Nat Genet. 1999;22(2):203–206. doi: 10.1038/9727. [DOI] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Hudson LD. Novel member of the zinc finger superfamily: a C2-HC finger that recognizes a glia-specific gene. Mol Cell Biol. 1992;12(12):5632–5639. doi: 10.1128/mcb.12.12.5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mall M, Kareta MS, Chanda S, Ahlenius H, Perotti N, Zhou B, et al. Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature. 2017;544(7649):245–249. doi: 10.1038/nature21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka S, Leon J, Tsuchimoto D, Sakumi K, Nakabeppu Y. MUTYH, an adenine DNA glycosylase, mediates p53 tumor suppression via PARP-dependent cell death. Oncogenesis. 2014;3:e121. doi: 10.1038/oncsis.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HM, Hu Z, Ma H, Greeley GH, Jr, Wang C, Englander EW. Developmental changes in expression and subcellular localization of the DNA repair glycosylase, MYH, in the rat brain. J Neurochem. 2004;88(2):394–400. doi: 10.1046/j.1471-4159.2003.02164.x. [DOI] [PubMed] [Google Scholar]
- McCarthy M. Allen Brain Atlas maps 21,000 genes of the mouse brain. Lancet Neurol. 2006;5(11):907–908. doi: 10.1016/s1474-4422(06)70594-4. [DOI] [PubMed] [Google Scholar]
- Errea O, Moreno B, Gonzalez-Franquesa A, Garcia-Roves PM, Villoslada P. The disruption of mitochondrial axonal transport is an early event in neuroinflammation. J Neuroinflamm. 2015;12:152. doi: 10.1186/s12974-015-0375-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130(3):548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22(1):103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Consalez GG, Hawkes R. The compartmental restriction of cerebellar interneurons. Front Neural Circ. 2012;6:123. doi: 10.3389/fncir.2012.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey MF, Peterson CL, Smale ST. Protein complex binding to promoter DNA: immobilized template assay. Cold Spring Harb Protoc. 2010;2010(8) doi: 10.1101/pdb.prot5465. pdb prot5465. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.