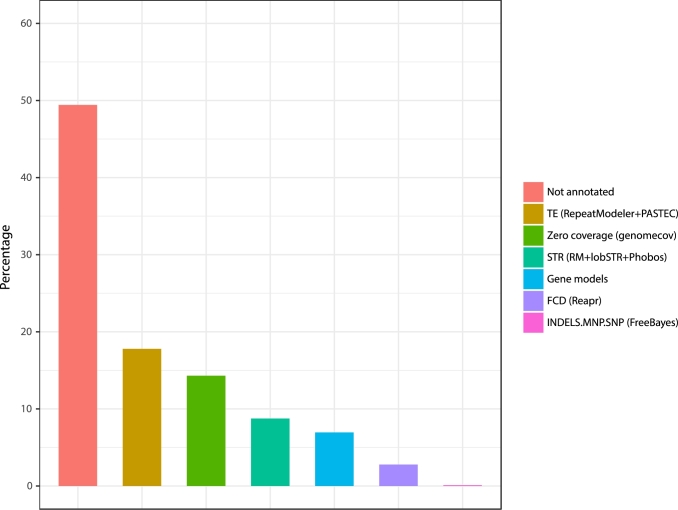
Figure 4:
Contig termini analysis to investigate the possible genomic features associated with gaps in the genome assembly. Contigs were created from the genome assembly with the “cutN -n 1” command from the seqtk program, which cut at each gap (of at least 1 base pair, i.e., 1 or more Ns). The figure shows the percentage of contig termini (the position of the terminal nucleotides of each contig) intersecting with different annotations of the genome.