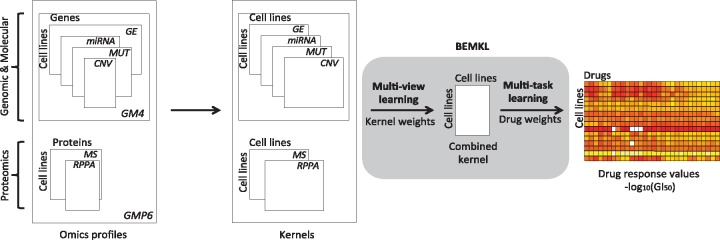
Fig. 1.
Data modeling approach, employing BEMKL method, applied on NCI60 genomics (point mutations and CNV), molecular (gene and miRNA expression) and proteomics (MS and RPPA) profiles across 58 pan-cancer cell lines to predict drug response of selected drugs. The BEMKL method learns the multi-view kernel weights to form a joint kernelized representation of the data which is used with the multi-task drug weights to model the response profile across the cell lines to each individual drug (the outcome matrix at the right)