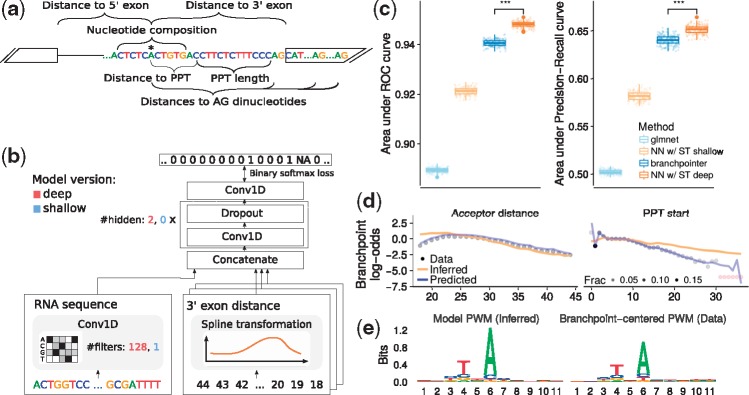
Fig. 3.
Spline transformation improves branchpoint prediction: (a) Features for branchpoint prediction designed by Signal et al. (2016) (adapted from Signal et al., 2016). (b) NN model architectures (deep and shallow) for branchpoint prediction developed here. For each predicted binary class (1 = high-confidence branchpoint, NA = ignored low-confidence branchpoint, 0 = else), the model takes as input 11 nt sequence window and 9 position-related features. (c) auROC and auPR bootstrap distribution (n = 200) for branchpoint prediction on the test set. Our NN models, spline transformation shallow (NN w/ST shallow) and spline transformation deep (NN w/ST deep), are compared to the state-of-the-art model branchpointer (Signal et al., 2016) and an elastic-net baseline using the same features. *** denotes P < 0.001 (Wilcoxon test). (d) Fraction of branchpoints per position for the two most important features in the log-odds scale (black dot, outlier shown in red) compared to the shallow NN model fit: inferred spline transformation (orange) and predicted branchpoint log-odds (blue). (e) Information content of the shallow NN convolutional filter transformed to the PWM and the branchpoint-centered PWM (Section 2)