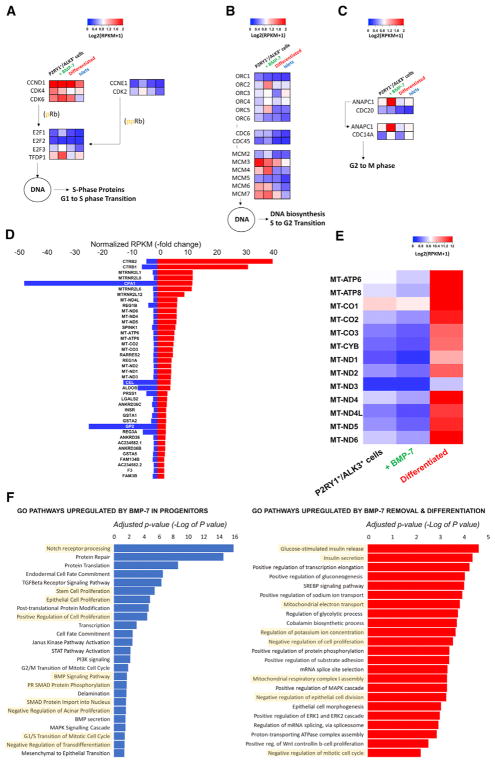
Figure 6. Analysis of BMP-7-Elicited Cell Replication and Differentiation Induced following BMP-7 Withdrawal.
(A–C) Key regulators of cell-cycle progression shown with their standardized log2 expression values across all four cell types. BMP-7 induces an upregulation of replication genes at all stages of cell division. In contrast, BMP-7 withdrawal (differentiation) leads to the overall downregulation of the replicative machinery.
(A) High cyclin D, CDK4, and CDK6 transcription is needed during the G1-to-S transition.
(B) Genes involved in the pre-replication Cdc45-Mcm2–7.CDC6 complex are recruited to the hetero-hexameric origin recognition complex (ORC2–6) and function as a replicative helicase unwinding pre-replicative DNA.
(C) The spindle assembly checkpoint is suppressed by the ANAPC1-CDC20 (APC/C) complex, allowing the G2-M transition (anaphase). Genes were chosen based on the KEGG Orthology reference hierarchy (KEGG: ko04110) and KEGG pathway entry (KEGG: map04110).
(D) BMP7-mediated gene expression down-regulation is inverted in differentiated cells. Fold changes in RPKM are shown for BMP7 treated versus P2RY1+/ALK3bright+ cells (blue) and differentiated versus P2RY1+/ALK3bright+ (red). The chart shows the top 40 coding genes with a fold change > 1 for the first comparison (blue bars) and >1.2 for the second (red bars). Only genes with a false discovery rate (FDR) <10% were considered.
(E) Heatmap showing gene expression across the three tissue types for mitochondrial protein-coding genes. (n = 3 biological replicates).
(F) Gene ontology (GO) analysis of P2RY1+/ ALK3bright cells treated with BMP-7 versus sorted P2RY1+/ALK3bright cells (left). GO analysis of differentiated cells versus BMP-7-treated (expanded progenitors) P2RY1+/ALK3bright cells (right).
n = 3 biological replicates.