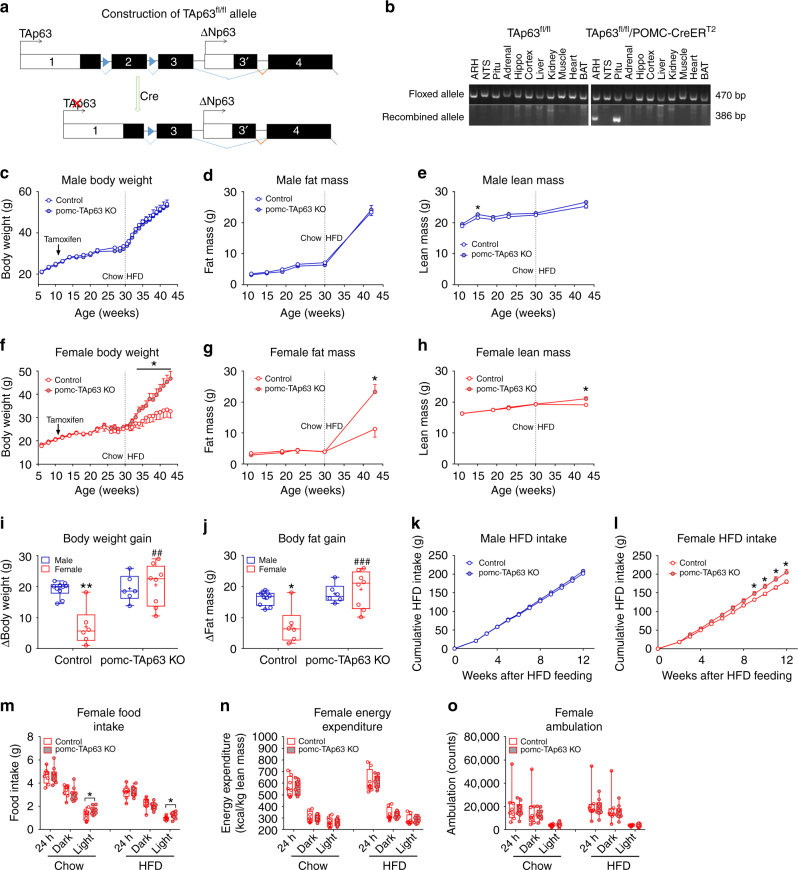
Fig. 3.
Deletion of TAp63 in POMC neurons diminishes sexual dimorphism in body weight responses to chronic HFD feeding. a Construction of TApfl/fl mouse allele. Two loxP sites flank exon 2 of the p63 gene and Cre-mediated recombination results in disruption of TAp63 expression; ΔNp63 expression starts from exon 3 and is therefore not affected by the recombination. b PCR detection of TApfl/fl allele and Cre-induced recombination in various tissues from the control (TAp63fl/fl) or the pomc-TAp63 KO (TAp63fl/fl/POMC-CreERT2) mice. Adrenal, adrenal gland; ARH, arcuate nucleus of the hypothalamus; BAT, brown adipose tissue; hippo, hippocampus; NTS, nucleus of the solitary tract; pitu, pituitary. c–e Body weight (c), fat mass (d), and lean mass (e) of male mice. Data are presented as mean ± SEM. N = 6 or 10 per group. *P < 0.05 in t-tests. f–h Body weight (f), fat mass (g), and lean mass (h) of female mice. Data are presented as mean ± SEM. N = 6 or 8 per group. *P < 0.05 in two-way ANOVA followed by post hoc Sidak tests. i–j Gains in body weight (i) and fat mass (j) during the 13-week HFD feeding. Data are presented as box and wiskers showing minimal, maximal, and median values with individual data points. N = 6–10 per group. *P < 0.05 or **P < 0.01 between male vs. female with the same genotype; ##P < 0.01 or ###P < 0.001 between the two genotypes within the same sex in two-way ANOVA followed by post hoc Sidak tests. k–l Cumulative HFD intake in male (k) and female (l) mice during the 13-week HFD feeding. Data are presented as mean ± SEM. N = 6 or 10 per group. *P < 0.05 in two-way ANOVA followed by post hoc Sidak tests. m–o Female control and pomc-TAp63 KO littermates (30 weeks of age) were adapted into the CLAMS metabolic cages and subjected to a 3-day chow 3-day HFD feeding protocol. Food intake (m), energy expenditure (normalized to lean mass, (n)), and ambulation (o) when fed chow or HFD. Data are presented as box and wiskers showing minimal, maximal, and median values with individual data points. N = 8 or 10 per group. *P < 0.05 between the two genotypes in t-tests