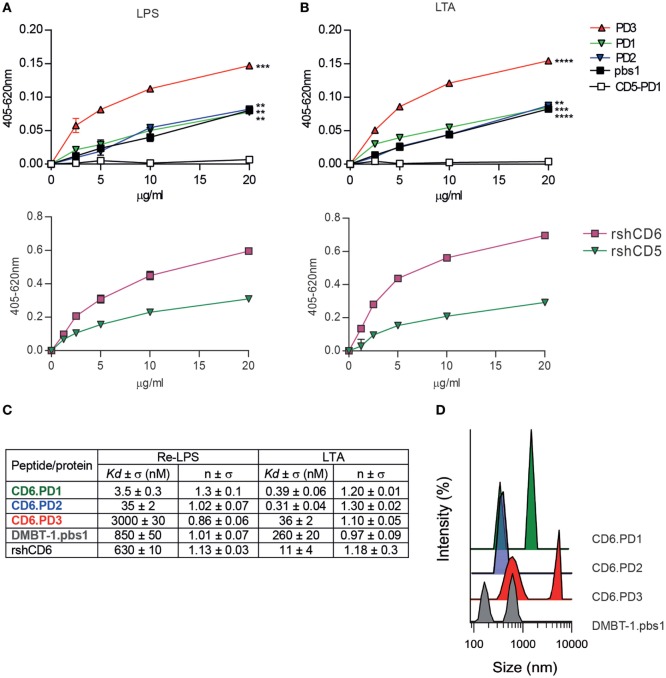
Figure 3.
Analysis of the direct binding characteristics of CD6-derived peptides to purified pathogen-associated molecular patterns (PAMPs) from Gram-negative and Gram-positive origin. Increasing concentrations (5–20 µg/mL) of biotin-labeled CD6 (PD1, PD2, and PD3), deleted in malignant brain tumors-1 (DMBT-1)/SAG- (pbs1), and CD5- (PD1) derived peptides (top panel), or rshCD5 and recombinant soluble human CD6 ectodomain (rshCD6) proteins (bottom panel) were added to 96-well ELISA plates sensitized with lipopolysaccharide (LPS) (A) or lipoteichoic acid (LTA) (B). Following overnight incubation at 4°C bound peptides or proteins were developed by addition of horseradish peroxidase–streptavidin and 3,3’,5,5’-tetramethylbenzidine substrate and further readings at OD 405–620 nm. Results are expressed as mean ± SD of duplicates from one representative experiment of three performed. Statistical analysis was performed by two-way ANOVA (*P < 0.05; **P < 0.01; and ***P < 0.001). (C) Apparent Kd values and Hill coefficients for the binding of peptides and proteins in study to LPS and LTA determined by tryptophan fluorescence. Peptides and proteins (10 µg/mL) were titrated with or without increasing concentrations of LPS or LTA in phosphate buffered saline (PBS). Results are mean ± SD of three experiments. (D) Dynamic light scattering analysis of the hydrodynamic diameter of CD6 (PD1, PD2, and PD3) and DMBT-1/SAG (pbs1)-derived peptides (10 µg/mL) in PBS. The y axis represents the relative intensity of the scattered light; the x axis denotes the hydrodynamic diameter of the particles present in the solution. One representative experiment of four is shown.