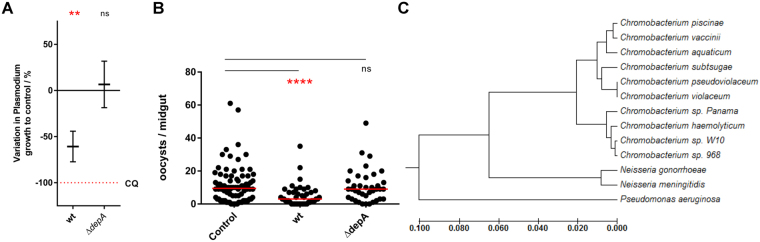
Figure 4.
Romidepsin production is necessary and sufficient for the antiplasmodial activity of Chromobacterium spp. (A) Variation in growth of asexual stage P. falciparum NF54 upon incubation with n-butanol extracts of approximately 5 mL of culture supernatants (LB, 72 h, 30 °C) of C. sp. 968 wildtype (wt) and depA-null mutant (∆depA). 0% inhibition is adjusted for parasite growth in vehicle control (1% DMSO) and 100% inhibition is matched to that of 250 nM chloroquine. Results are shown as mean ± standard deviation; significance was determined using a one-tailed one sample t-test to determine whether each treatment significantly lowered parasite growth compared to control (ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001). CQ, chloroquine. (B) Female An. gambiae were provided either PBS, C. sp. 968 wildtype (wt) or its ∆depA mutant suspended within 3% sucrose. After 1 day of being allowed to feed on these suspensions, they were given a P. falciparum NF54-infected blood meal, and oocyst numbers were determined 7 days later; oocyst counts for three independent replicates are shown. Horizontal bars represent the median number of oocysts per treatment; inhibition (%) was estimated based on the comparison of these values to that of the PBS control. Prevalence represents the proportion of infected mosquitoes per group. Significance was determined using the Kruskal-Wallis test by comparing treatment groups to the control and correcting for multiple comparisons by the Dunn’s test (ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001). (C) Evolutionary relationships between C. sp. 968, other chromobacteria and other related species (Neisseria gonorrhoeae NCTC13800; Neisseria meningitidis LNP21362; Pseudomonas aeruginosa DSM 50071) based on 16 S rRNA genomic sequences as inferred by the UPGMA method. The optimal tree with the sum of branch lengths equal to 0.33018119 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis was conducted in MEGA6 and involved 13 nucleotide sequences for a total of 925 positions in the final dataset.