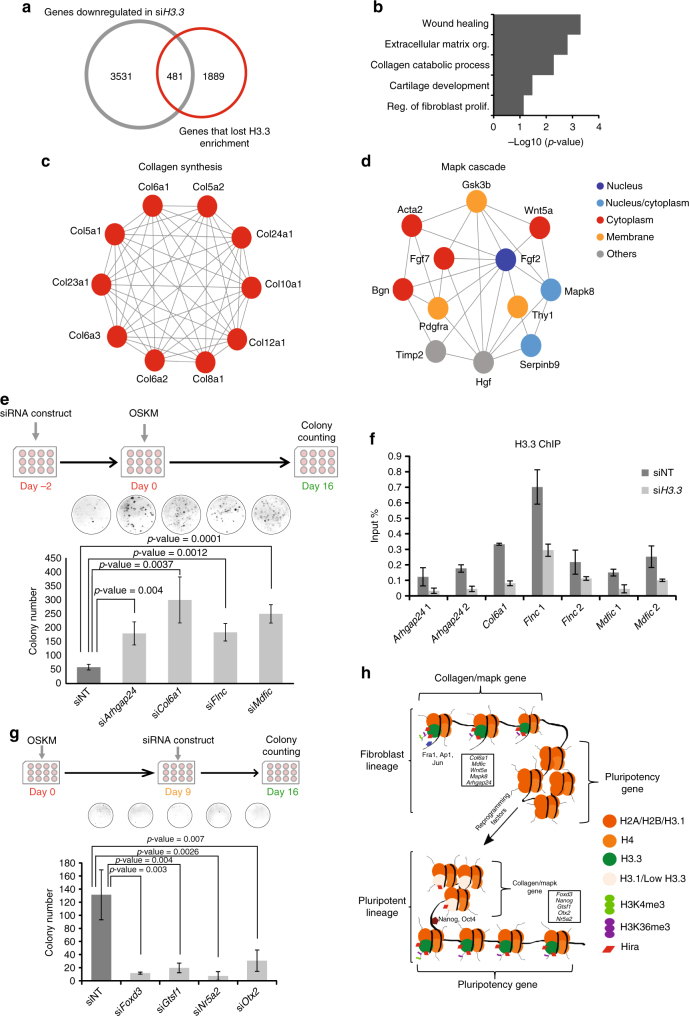
Fig. 7.
H3.3 regulates processes associated with fibroblast lineage. a Venn diagram correlating genes with reduced expression upon knockdown of H3.3 and genes with reduced H3.3 enrichment as cells progress via the successful route of cellular reprogramming. b GO analysis of the functional targets of H3.3. X-axis represents the GO term enrichment score. c, d Protein–protein interaction network of functional H3.3 targets associated with collagen synthesis (c) and Mapk cascade pathway (d). e Schematics of the knockdown experiment (top). The bar chart below represents the number of colonies (y-axis) observed in wells in which the knockdown of the indicated genes had been performed. Non-targeting siRNA constructs were used as controls (siNT). The images above the bar charts are representative images of the wells in which the counting took place. Values are mean ± s.e.m from independent replicate experiments (n = 3). Two-tailed t-test was used for statistical analysis. Error bars represent standard deviation. f Bar chart demonstrating the level of H3.3 enrichment on the genes indicated in the x-axis upon the knockdown of H3.3 (siH3.3). Non-targeting siRNA (siNT) were used as controls. Y-axis represents the enrichment level in the ChIP sample over input. Error bars represent standard deviation (n = 3). g Schematics of the knockdown experiment (top). The bar chart below represents the number of colonies (y-axis) observed in wells in which the knockdown of the indicated genes had been performed. Non-targeting siRNA constructs were used as controls (siNT). The images above the bar charts are representative images of the wells in which the counting took place. Values are mean ± s.e.m from independent replicate experiments (n = 3). Two-tailed t-test was used for statistical analysis. Error bars represent standard deviation. h Dynamic deposition of H3.3 during cellular reprogramming. H3.3 is enriched at genes associated with Mapk cascade or collagen synthesis where it cooperates with the AP1–Fra1 complex to safeguard the fibroblast identity. During reprogramming, H3.3 are disassembled from the lineage genes. Reconfiguration of H3.3 to the nucleosomes of the reprogrammed pluripotent lineage ensure the establishment of the new fate. The effect of H3.3 is exerted via its deposition by Hira and the methylation of its K4 and K36 residues