Fig. 7.
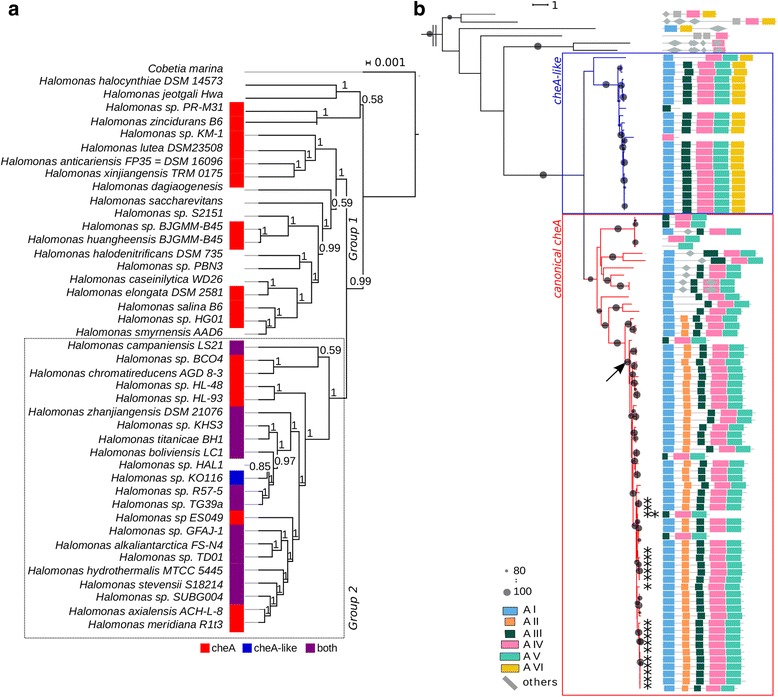
CheA and CheA-like protein phylogeny largely follows taxonomy-based topology. a 16S + 23S Halomonas sp. Phylogenetic Tree. 16S and 23S data were retrieved from the available Halomonas sp. genomes and aligned with SINA, specifying SSU or LSU, respectively. Both alignments were attached and phylogenetic analyses were performed with BEAST package. The colored squares indicate the presence of canonical cheA sequence only (red), cheA-like sequence only (blue) or both types (purple); no colored squares indicate lack of both cheAs in those genomes. This tree shows a main bifurcation that largely explains the canonical CheA / CheA-like clade separation (dot line square enclosing the group 2 where Halomonas strains with two clusters are included). b CheA protein phylogenetic analyses. In red, canonical CheA clade. In blue, CheA-like clade. Protein domains are depicted for each sequence. The black arrow indicates the root of the Halomonas sp. CheA clade. Asterisks indicate strains that have the two clusters. Big gray circles represent bootstrap support 100%; small gray circles represent bootstrap support 80%. The two lines at the start of the tree indicate where it was trimmed. The complete unrooted tree can be seen in Additional file 5: Figure S3
