FIG 1 .
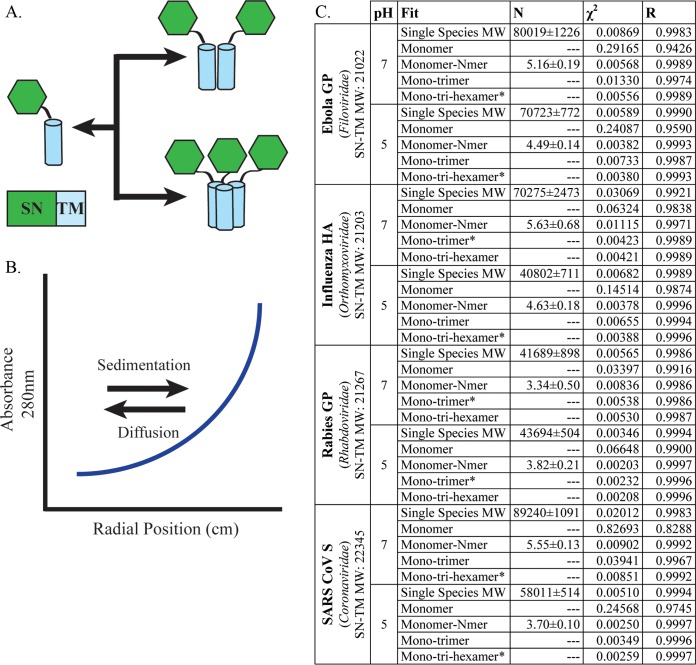
Parameters for SE-AUC. (A) Chimeric proteins were designed for SE-AUC, in which the TMD of interest was fused with the protein staphylococcal nuclease (SN). The addition of SN aids in purification and increases the extinction coefficient, which aids data collection in absorbance-based assays. (B) SE-AUC is based on the premise that, upon centrifugation, the species present will eventually reach an equilibrium wherein species of higher mass (such as a higher oligomeric state) will sediment, while species of lower mass will diffuse. The resulting spectra represent an equilibrium in which higher-molecular-weight species have a higher absorbance. The absorbance data can be fit using the following equation: , where A represents the total absorbance of the solution at radial position (r), while αm,0 and αm/t,0 represent the monomer (m) and monomer/trimer (m/t) absorbance, respectively, as the reference radius, r0. The molecular mass (Mm) and partial specific volume (v) of the monomer in solution were estimated using SEDNTERP (http://www.rasmb.org/sednterp/). The molecular mass for a monomer or trimer (Md/t) is a multiple of Mm. R is the universal gas constant; T is the absolute temperature; ρ is the solvent density; ω is the angular velocity; and ξ is the baseline offset. With the known molecular weight of the single species, the oligomeric state can be determined. The program KaleidaGraph was used to fit these equations to the data. (C) Table includes the viral fusion protein, its family, and χ2 and R values for the curve fits shown in Fig. 2 at both pH 7 and 5. The single-species molecular weight (MW) is indicated for each SN-TM construct, and the best fit is indicated by an asterisk.