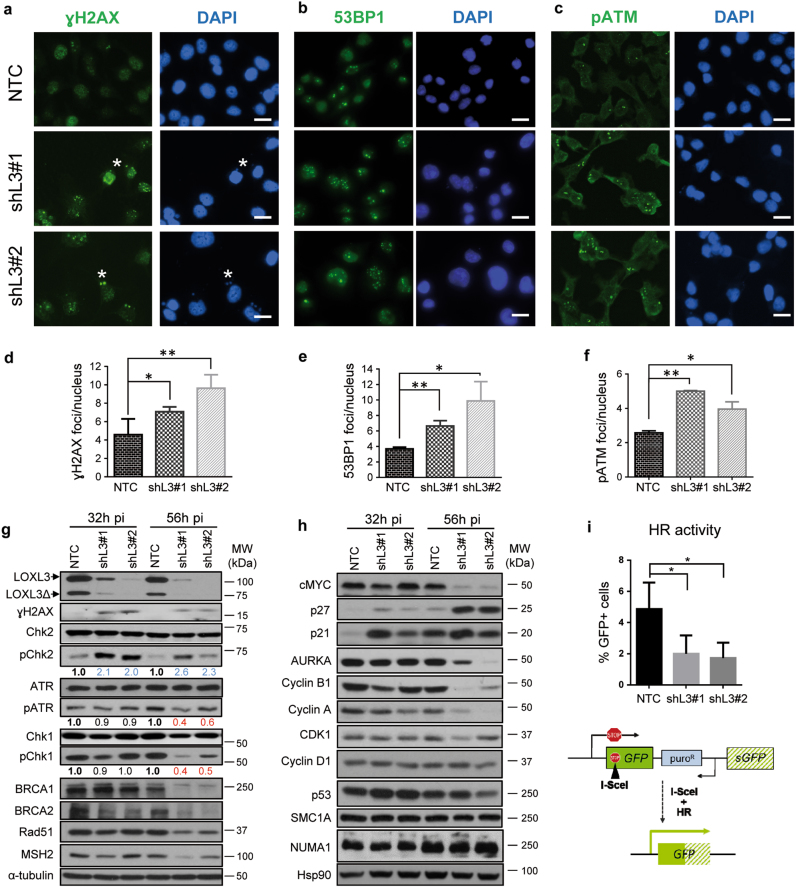
Fig. 5. LOXL3 is involved in the DNA damage checkpoint.
(a–c) A375P cells were transduced with control (NTC) and LOXL3 targeting (shL3#1 and shL3#2) lentivirus and collected at 56 h pi, fixed and processed for γH2AX (a), 53BP1 (b), and phospho-ATM (pATM) (c) immunofluorescence. DAPI (blue) was used to stain DNA. Representative micrographs are shown. White asterisks a mark the presence of γH2AX micronuclei. Scale bar: 20 μm. (d-f) Quantitation of γH2AX (d), 53BP1 (e) and pATM (f) immunofluorescence foci. Error bars represent s.e.m., n = 5 biologically independent replicates for γH2AX (1000 total cells analyzed), n = three biological replicates for 53BP1 (450 total cells analyzed) and n = 2 biological replicates for pATM (500 total cells analyzed). ∗p < 0.05, ∗∗p < 0.01 by a two-sided Student’s t-test. (g and h) Whole-protein extracts from control (NTC) and LOXL3-silenced (shL3#1 and shL3#2) A375P cells after 32 and 56 h pi were subjected to western blot analyses with the indicated antibodies. AURKA: Aurora Kinase A. α-tubulin (g) and Hsp90 (h) were used as loading controls. The phospho-ATR (pATR), phospho-Chk1 (pChk1), and phospho-Chk2 (pChk2) levels were calculated normalizing to the levels of the corresponding total proteins using ImageJ and their relative levels regarding NTC control cells are indicated below the matching immunoblots. Data shown are representative of three biologically independent replicates. MW of protein standards (kDa) is indicated (g, h). (i) HR activity represented as the percentage of GFP-positive cells analyzed by FACS from A375P pHPRT-DRGFP cells transduced with control (NTC) and LOXL3 shRNA lentivirus (shL3#1 and shL3#2) after I-SceI transfection. Error bars represent s.d., n = 4 biologically independent experiments. ∗p < 0.05 by a two-sided Student’s t-test. Lower panel displays a scheme summarizing the experiment performed. pHPRT-DRGFP reporter consists of two defective GFP genes, the first contains an I-SceI endonuclease site. Subsequent cellular expression of I-SceI leads to a DSB that can be repaired by HR using the downstream wild-type GFP sequence as a template, resulting in GFP-positive cells