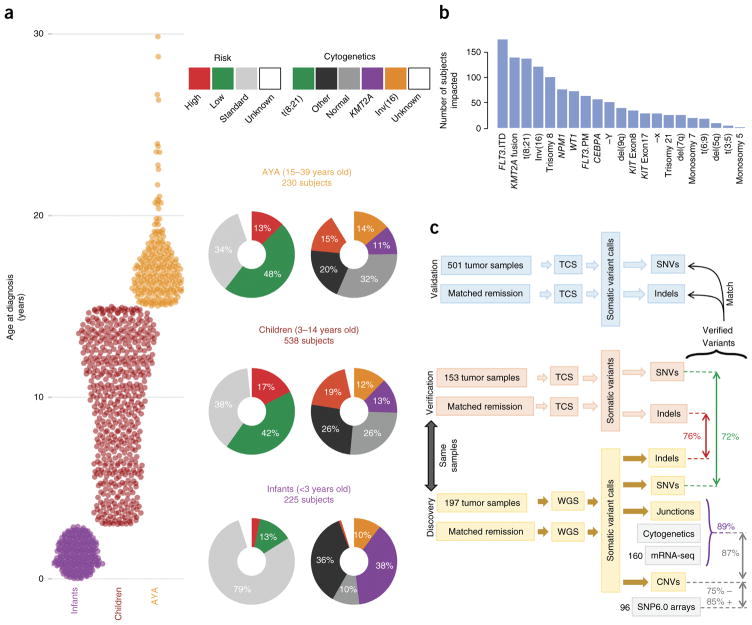
Figure 1.
An overview of the TARGET AML study. (a) The distribution of subjects by clinical risk category and cytogenetic classification is shown adjacent to each age group analyzed (infant, <3 years old; child, 3–14 years old; AYA, 15–39 years old). KMT2A indicates cytogenetic KMT2A abnormality (t(11q23)). (b) Summary of the clinically established molecular aberrations in the cohort (n = 993 subjects). FLT3.ITD, FLT3 internal tandem duplications; FLT3.PM, FLT3 Asp835 point mutations; KIT Exon8 and KIT Exon17, KIT mutations impacting the named exons. (c) Overview of the genomic variant discovery, verification and validation process. We characterized diagnostic and remission (taken as germline) samples from 197 subjects using WGS and verified 153 diagnostic–remission case pairs using TCS of genes recurrently impacted in the WGS samples (an additional 29 WGS cases were verified by TCS of diagnostic cases only; Supplementary Fig. 1). Seventy-two percent of WGS SNVs and 76% of WGS indels were confirmed through TCS (red and green text, respectively). Purple text indicates the percentage of confirmed DNA junctions. For focal copy number (CN) alterations spanning fewer than seven genes, 75% of recurrent WGS deletion or loss and 85% of gain or amplification calls matched recurrent alterations discovered by SNP6 array in 96 matching samples. For chromosomal junctions, we integrated WGS, clinical karyotyping and RNA-seq data by majority vote, confirming 89% of WGS junction calls.