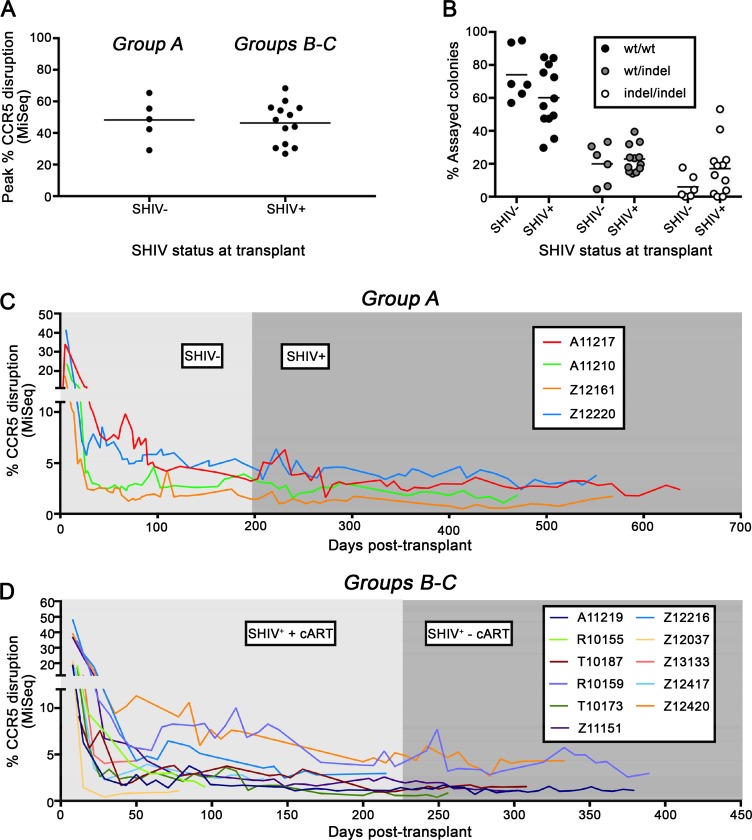
Fig 2. ΔCCR5 cells persist in peripheral blood of SHIV+ animals.
Following isolation of autologous HSPC from SHIV-naïve (Group A) and SHIV-infected animals (Groups B-C) and ex vivo CCR5-gene editing, deep sequencing was used to quantify the percentage of edited CCR5 alleles in the HSPC infusion product and in transplanted animals. (A): Peak CCR5 disruption in CD34+ HSPC infusion products cultured for 5–12 days ex vivo. (B): Colony assay quantifying the percentage of wt, mono-, and bi-allelically disrupted, CD34+ HSPC-derived colonies. Error bars represent standard error of the mean for the percentages of at least 48 colonies derived from 5 SHIV- and 12 SHIV+ animals. (C): CCR5 gene editing in peripheral blood samples from previously SHIV-naïve animals before and after SHIV infection. (D): CCR5 gene editing in peripheral blood samples from previously SHIV-infected and cART-suppressed animals before and after release of cART.