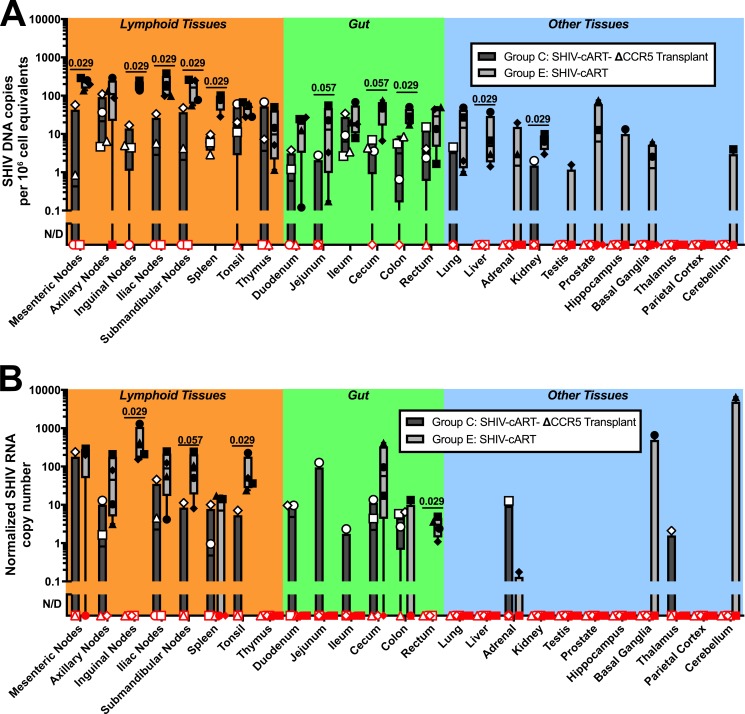
Fig 8. ΔCCR5 transplant decreases tissue viral reservoirs in stably suppressed animals.
Following SHIV infection and cART suppression, necropsy and extensive tissue collections were performed on Group E animals (n = 4) that were not transplanted (“SHIV-cART,” light gray bars + closed circles) and Group C animals (n = 4) that were transplanted with CCR5 gene-edited cells (“SHIV-cART-ΔCCR5 Transplant,” dark gray bars + open shapes). All animals were stably suppressed at the time of necropsy, when tissues were collected from the indicated sites, and nucleic acids were extracted from total tissue homogenates. (A) SHIV DNA copies per 106 cell equivalents. (B) Normalized SHIV RNA copy number. Quantitative PCR was used to measure SHIV DNA and RNA, and normalized to macaque RNase P p30 (MRPP30) DNA and RNA, respectively. Red shapes indicate samples in which SHIV DNA or RNA was not detected (ND) despite 4 (DNA) or 2 (RNA) re-tests of the original negative sample. Positive values represent the quantity on initial testing, or for samples undergoing repeat testing, the average of all results. Cell input for each assay as determined by MRPP30 is shown in S2 Table. Shapes indicate individual values for one animal. Animal IDs for open shapes: circles, Z12351; squares, Z13133; triangles, Z12216; diamonds, Z12417. Animal IDs for closed shapes: circles, A11213; squares, A11221; triangles, A11197; diamonds, A11198. Bars indicate maximum/minimum values. Exact p-values for significant (p < 0.05) and near-significant differences are indicated.