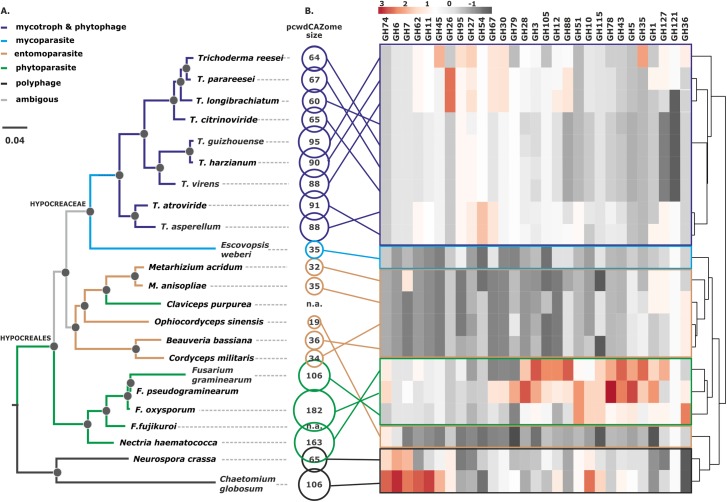
Fig 1. Phylogeny of Hypocreales and the composition of their pcwdCAZomes.
A. Bayesian phylogram obtained based on the curated concatenated alignments of 100 orthologous neutrally evolving proteins of Hypocreales and two other Sordariomycetes. Black dots above nodes indicate posterior probability support > 0.95. The colors of the branches indicate the major nutritional strategy in the group (see insert) as described in Supporting Information S1 Text. B. The size of each pcwdCAZome per species is shown as a circle; n.a. means not available. The heat map shows the gene number for each GH family in the Hypocreales fungi examined; cluster analysis was performed with Euclidian distance and complete linkage for rows. The corresponding data matrix is presented in Supporting Information S3 Table. GH indicates glycosyl hydrolase family.