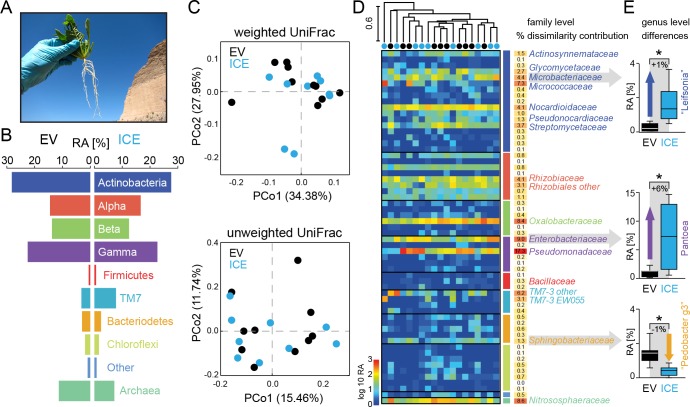
Figure 6. The overall composition of the root-associated microbial communities from field-grown plants did not differ between genotypes and showed marginal effects at the genus level.
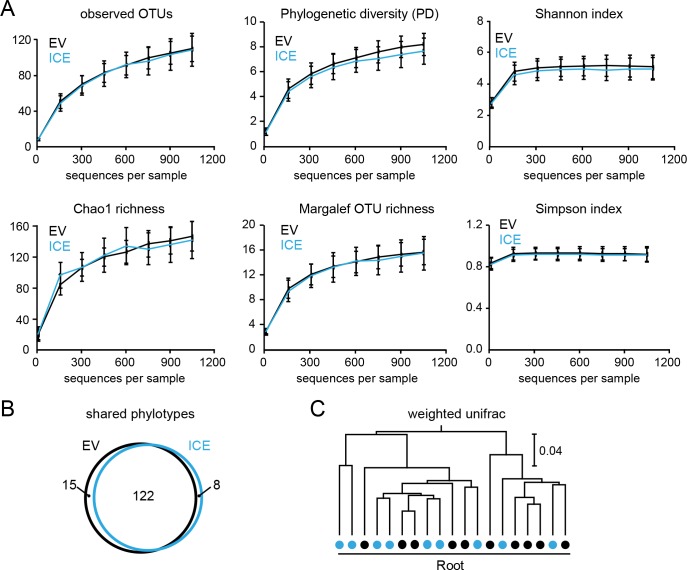
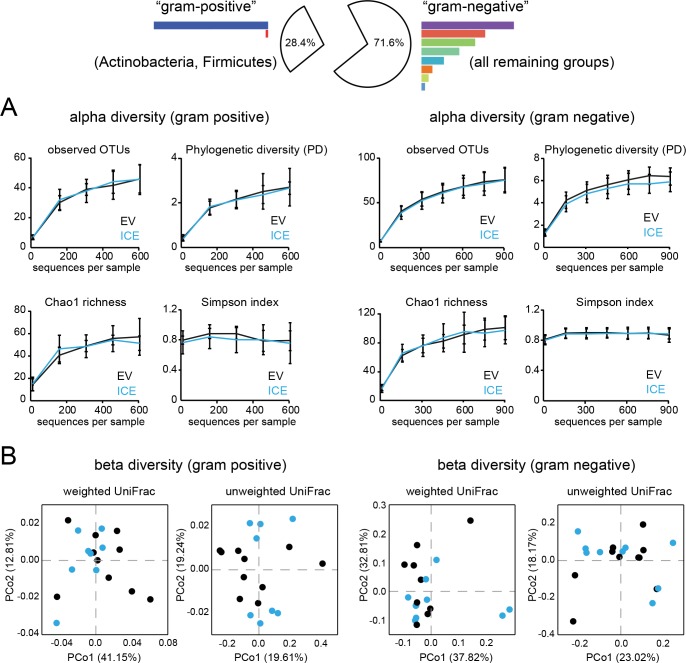
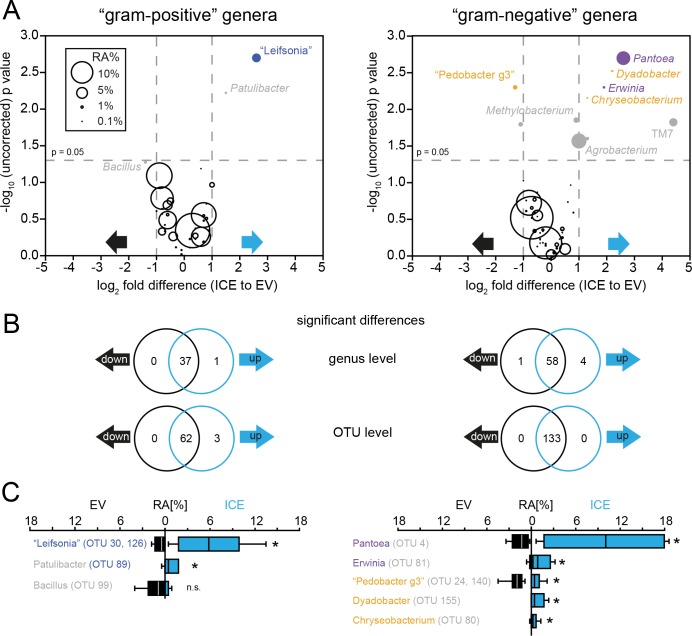
(A) Root samples from ten equal sized plant pairs of empty vector control (EV) and ICE 8 plants were used for sequencing. (B) Comparison of the relative abundances of the major phyla (respective classes for Protobacteria) of the root-associated microbial communities of field-grown plants. (C) Communities were clustered by principal coordinates analysis (PCoA) using the weighted and unweighted UniFrac as a distance measure (genotype: EV = black, ICE 8 = blue). (D) The filtered community data was used to visualize the distribution of all 59 families in a heatmap showing log10 (+1) transformed abundance. Samples were clustered hierarchically by UPGMA based on Bray–Curtis dissimilarity. Similarity percentage analysis (SIMPER) was performed to evaluate sample dissimilarity contribution [%] for each family based on their relative abundance. Only families with >1% dissimilarity contribution are indicated by name. Color coding of the bars indicate higher phyla and classes as used in B. (E) Group significance tests revealed significant differences between control and antimicrobial peptide expressing plants at the genus level for ‘Leifsonia’ (Microbacteriaceae), Pantoea (Enterobacteriaceae) and ‘Pedobacter_g3’ (Sphingobacteraceae) (non-parametric t-test, *p<0.05 after correcting for a false discovery rate of 10%).