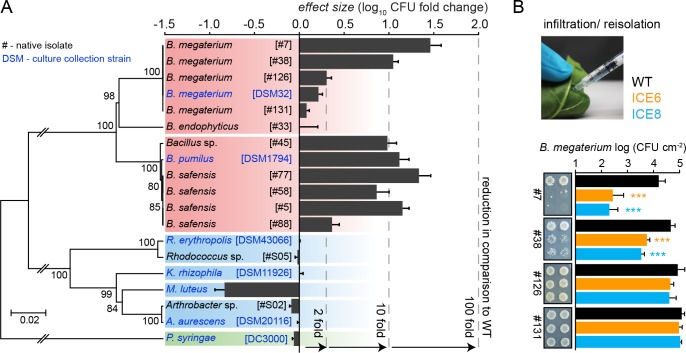
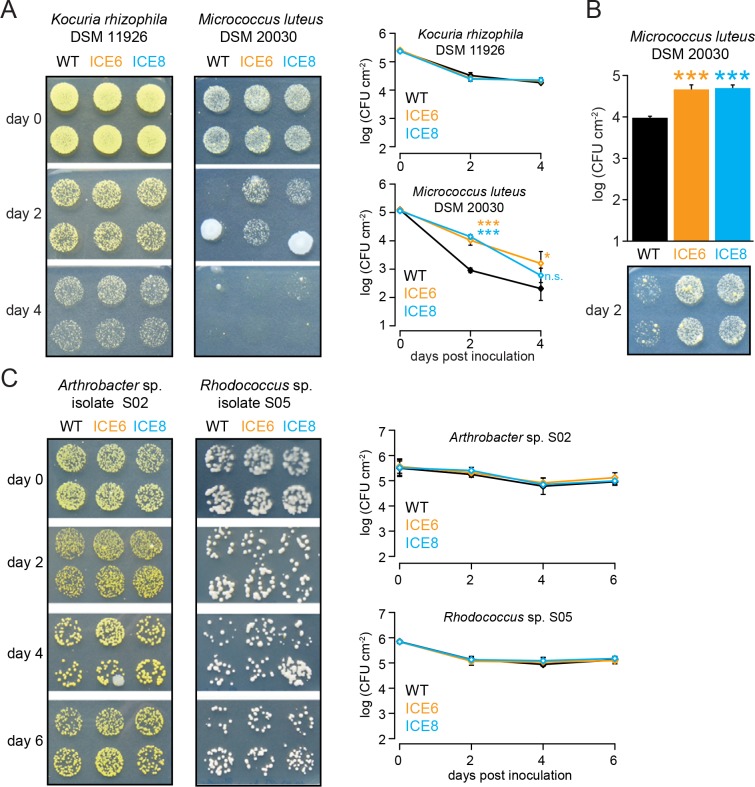
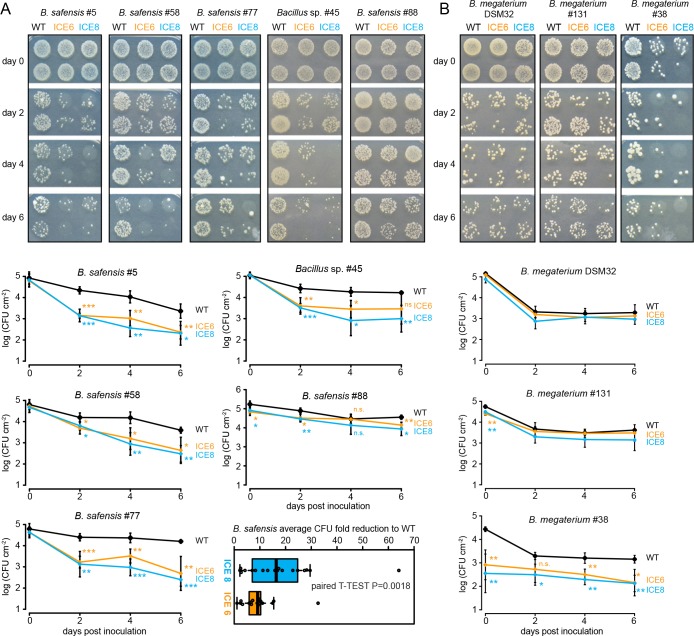
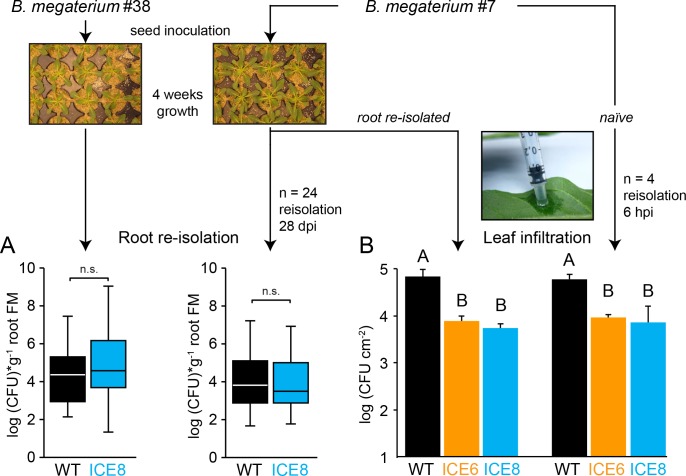
Figure 7. Experimental infiltrations of bacterial isolates from different taxa demonstrate the specificity of the antimicrobial activity.
(A) Summary of the in planta antimicrobial effects obtained by individual infiltrations of bacterial isolates into transgenic and control plants, as shown in Figure 7—figure supplement 1 and Figure 7—figure supplement 2. Bars represent the effect size (averaged log10 CFU fold reduction to WT) calculated by dividing the CFUs obtained from WT by the CFUs obtained from the transgenic plants (ICE 6 and ICE 8) from 2 to 6 dpi or 6 hpi (only B. megaterium). The relative phylogenetic grouping of the bacterial strains is illustrated by a neighbor-joining tree, generated from the alignment of the nearly complete 16S rDNA sequences (Figure 7—source data 1). Bootstrap supported values (1000 replicates) are indicated on individual nodes. The theoretical OTU clustering (97% similarity of the pyrosequencing amplicon) is indicated by the gaps in the background color (Firmicutes in red, Actinobacteria in blue and Proteobacteria in green). (B) Native isolates of Bacillus megaterium showed strain specific susceptibilities to the transgenic plants. Bacteria were injected in the leaves by pressure infiltration and re-isolated after 6 hr (±SD, n = 4 plants). Asterisks indicate statistically significant differences between WT and transgenic plants (students t-test; ***p≤0.001).