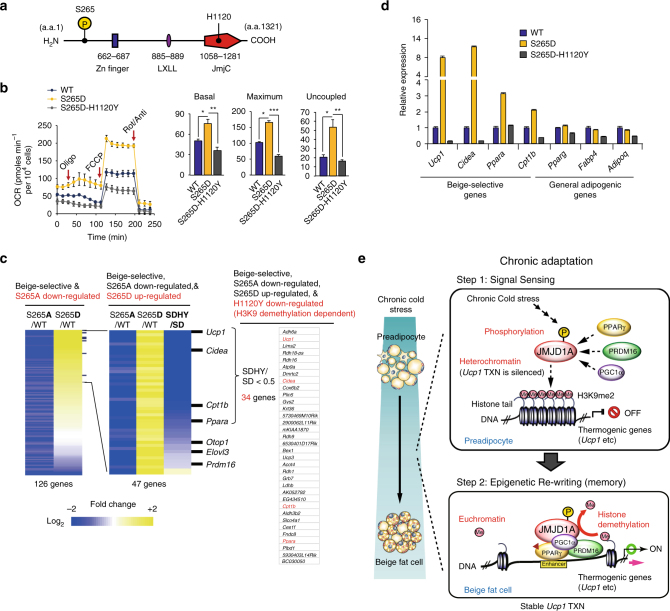
Fig. 6.
Demethylation activity is pivotal for beige-selective gene inductions. a Schematic representation of the domain structure of human JMJD1A. Phosphorylation site at S265 and Fe(II) binding site at H1120 are shown. b OCR in im-scWATs stably expressing WT-hJMJD1A, S265D-hJMJD1A, or S265D-H1120Y-hJMJD1A treated sequentially with oligomycin (Oligo), FCCP, and rotenone/antimycin A (Rot/Anti) (left). Basal, maximum, and uncoupled respiration calculated from the left (right). Data are mean ± s.e.m. of three technical replicates in a representative experiment. Analysis of variance were performed, followed by Tukey’s post hoc comparison. *P < 0.05, **P < 0.01, and ***P < 0.005 were considered statistically significant. c Heat map of mRNA levels determined by RNA-seq analysis of the indicated WT or mutant versions of hJMJD1A expressing im-scWATs. Changes are log2 expression (FPKM) ratios. For reference, a color intensity scale is included. Thirty-four genes that are beige-selective, S265A downregulated, S265D upregulated, and H1120Y downregulated (H3K9 demethylation-dependent) are listed (right). d qPCR analysis confirming RNA-seq shown in c. Mean ± s.e.m. of three technical replicates. e Hypothetical model. Cold stimulated β-adrenergic signal leads to phosphorylation of JMJD1A (Step 1), which triggers the formation of a PRDM16-PGC1α-PPARγ transcription complex that targets beige-selective genes (Step 1, top). JMJD1A then demethylates H3K9me2 to turn on the transcription of these genes in scWAT (Step 2, bottom)