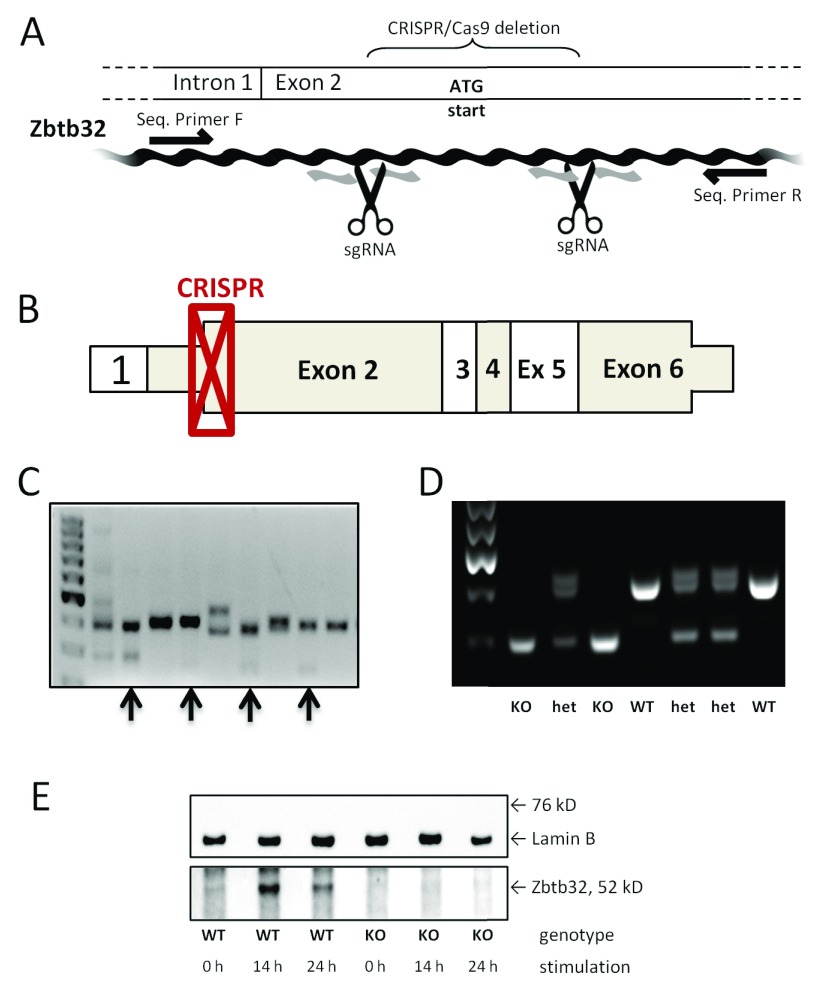
Figure 1. Confirmation of CRISPR-mediated knockout of Zbtb32.
The CRISPR/Cas9 technique was used to target exon 2 of the Zbtb32 gene directly in NOD mice. A portion of the Zbtb32 gene was deleted and a frameshift introduced in NOD mice using the CRISPR/Cas9 technique, as illustrated here ( A). The scissors represent the location of cutting based on sgRNA binding and black arrows indicate binding sites for the standard primers, which were used for both sequencing and genotyping. The location of the deletion maps onto the beginning of exon2 of the wildtype mRNA ( B). Non-coding portions are shown as thinner than the coding portions. The CRISPR/Cas9-treated embryos develop into mice with genetic mosaicism of the Zbtb32 gene in the F0 generation. Four genetically heterogeneous females (indicated with black arrows) were examined further before one mutant allele was chosen to establish the NOD.Zbtb32 -/- colony ( C). The targeted deletion of Zbtb32 removed 146 bp from exon 2 as shown by genetic screening of knockout, heterozygous, and wildtype animals in the F2 generation ( D). The loss of the Zbtb32 protein after CRISPR-mediated deletion was confirmed by Western blotting extracts from stimulated CD4 + splenocytes ( E) (from three biologically independent replicates).