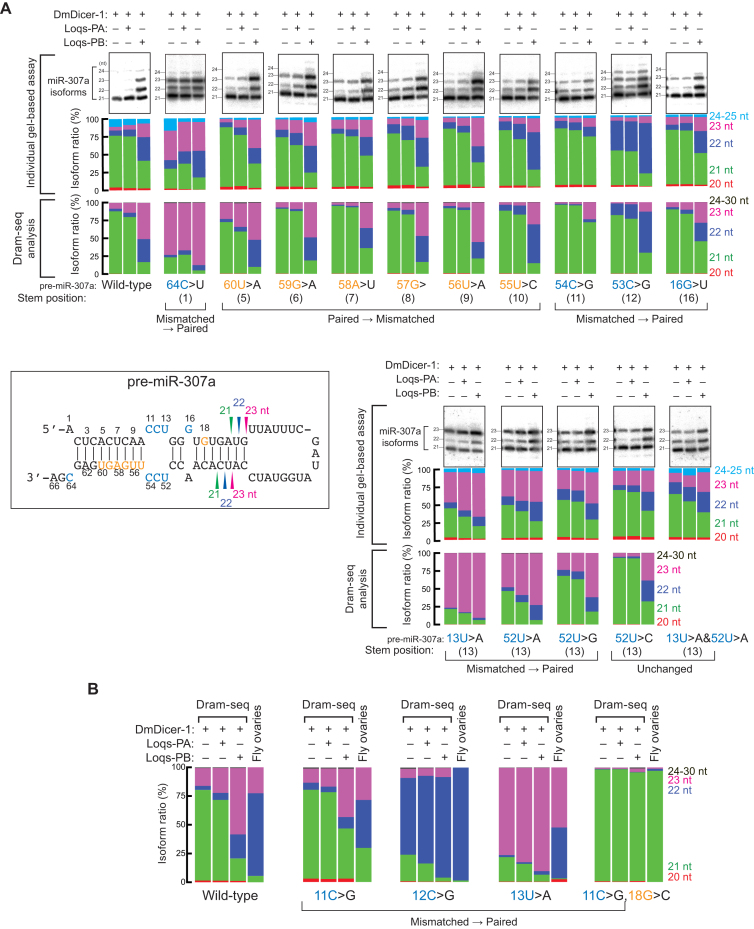
Figure 3.
miR-307a length distributions revealed by Dram-seq, individual gel-based in vitro assay, and sequencing from fly ovaries. (A) In vitro dicing of 100 nM pre-miR-307a variants by 8 nM DmDicer-1 ± Loqs-PA or Loqs-PB for 120 min. The miR-307a isoforms were detected by northern blot (top rows); length distributions revealed by quantification of gels (means of at least three independent trials) (middle rows); length distributions revealed by Dram-seq (bottom rows). (B) Length distributions of miR-307a isoforms produced from wild-type pre-miR-307a and its variants by recombinant DmDicer-1 ± Loqs-PA or Loqs-PB in vitro revealed by Dram-seq (left threes). Length distributions of miR-307a isoforms produced from the same pre-miR-307 variants in fly ovaries (right). Fly strains w1118; miR-307anull; UASP-miR-307a transgenes [wild-type and point mutants]/mat-15-Gal4.