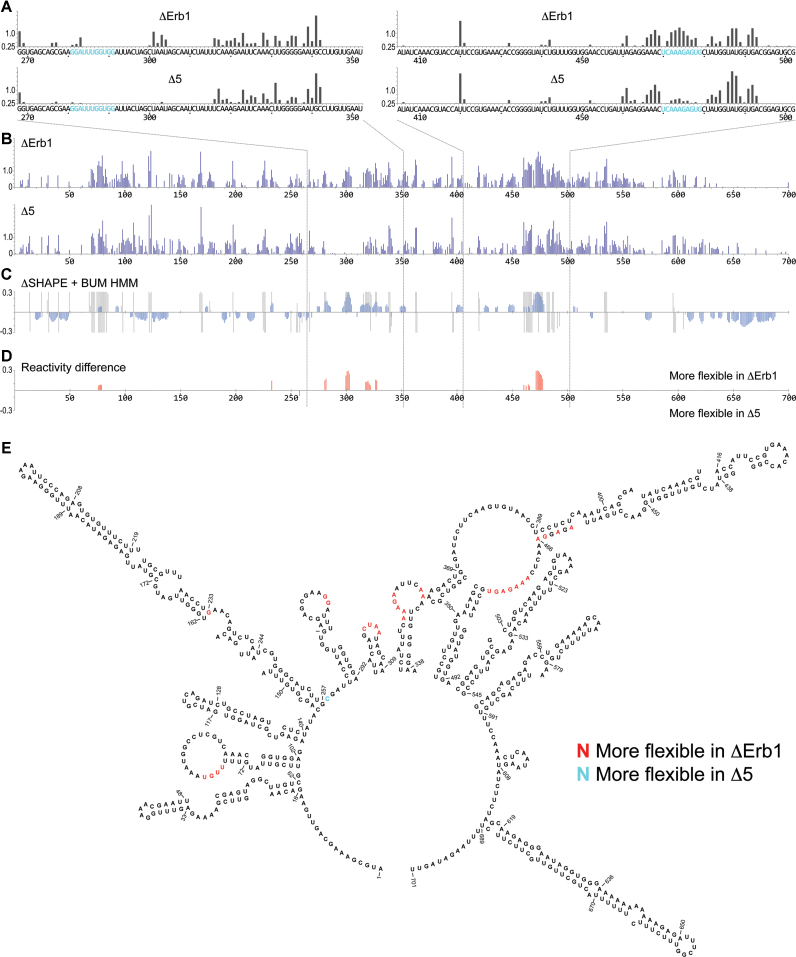
Figure 3.
SHAPE reactivity pattern for the 5′ETS region in 35S pre-rRNA. (A) Primer extension determined SHAPE reactivities of nucleotides 268–355 (left) and nucleotides 405–504 (right) in ΔErb1 and Δ5. U3 base pairing sites, proposed in (20–23), are shown in blue. (B) ChemModSeq analyses for the entire 5′ETS (nucleotides 1–700) of 35S pre-rRNA in ΔErb1 and Δ5. (C) Comparison of SHAPE reactivities in ΔErb1 and Δ5, using ΔSHAPE analyses and BUM-HMM probabilities. Blue bars show the ΔSHAPE values. Gray regions mark nucleotides with posterior probabilities of ≥0.95 according to BUM-HMM analyses for ΔErb1 (top) and Δ5 (bottom). (D) ΔSHAPE reactivity differences between ΔErb1 and Δ5 that overlap with the BUM-HMM probabilities. In red, more flexibility in ΔErb1, in blue, more flexibility in Δ5. The regions analysed by primer extension are indicated by dashed lines. (E) Reactivity differences marked in a secondary structure model of the 5′ETS (44). Red nucleotides were more reactive in ΔErb1, blue nucleotides were more reactive in Δ5.