Figure 1.
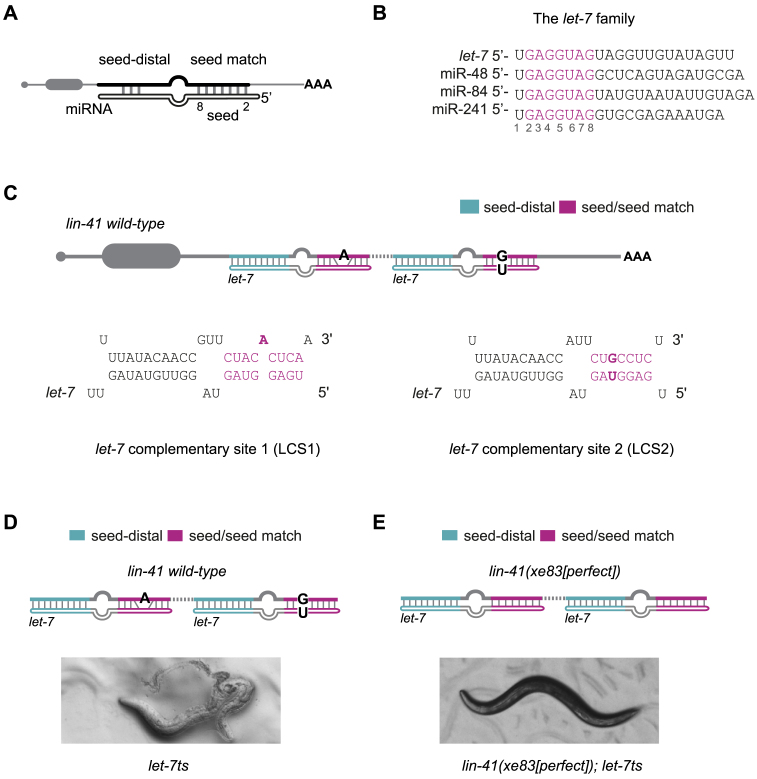
let-7 becomes dispensable for viability when the lin-41 3′UTR contains perfect seed match sites. (A) Schematic drawing of a miRNA/target duplex with seed (nucleotides 2–8)/seed match and limited seed-distal pairing indicated. Top mRNA, bottom miRNA. (B) The let-7 family with the seed sequence (nucleotides 2–8) highlighted in magenta. (C) The two let-7 complementary sites (LCS1 and LCS2) in the lin-41 3′UTR of C. elegans. Each site contains an imperfect seed match (a bulged A and a G: U wobble, respectively, in bold) to the let-7 family and an extensive seed-distal pairing to let-7 only. The sites are separated by 27 nt of intervening sequence (dashed line). (D, E) Representative images of animals carrying the let-7ts mutation and (D) wild-type lin-41 or (E) the lin-41(xe83[perfect]) allele with perfect seed match to the let-7 family and unchanged seed-distal region. Animals were grown at 25°C. let-7ts: let-7(n2853) X, temperature sensitive lesion. miRNA site legend: magenta = seed/seed match; cyan = let-7 seed-distal binding.